使用annovar软件参考自:http://www.bio-info-trainee.com/?p=641
/home/jmzeng/bio-soft/annovar/convert2annovar.pl -format vcf4 Sample3.varscan.snp.vcf > Sample3.annovar
/home/jmzeng/bio-soft/annovar/convert2annovar.pl -format vcf4 Sample4.varscan.snp.vcf > Sample4.annovar
/home/jmzeng/bio-soft/annovar/convert2annovar.pl -format vcf4 Sample5.varscan.snp.vcf > Sample5.annovar
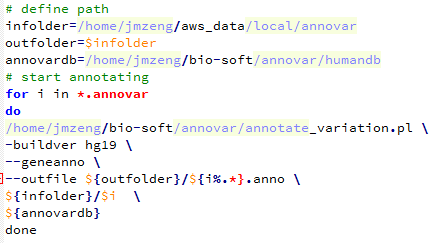
然后用下面这个脚本批量注释
Reading gene annotation from /home/jmzeng/bio-soft/annovar/humandb/hg19_refGene.txt ... Done with 50914 transcripts (including 11516 without coding sequence annotation) for 26271 unique genes
最后查看结果可知,真正在外显子上面的突变并不多
23515 Sample3.anno.exonic_variant_function
23913 Sample4.anno.exonic_variant_function
24009 Sample5.anno.exonic_variant_function
annovar软件就是把我们得到的十万多个snp分类了,看看这些snp分别是基因的哪些位置,是否引起蛋白突变
downstream
exonic
exonic;splicing
intergenic
intronic
ncRNA_exonic
ncRNA_intronic
ncRNA_splicing
ncRNA_UTR3
ncRNA_UTR5
splicing
upstream
upstream;downstream
UTR3
UTR5
UTR5;UTR3