这是一个在线工具,非常好用,对snp位点进行注释,看看该突变是否影响蛋白功能,一定要收藏!!!
官网:http://mutationassessor.org/
也应该是有standalone版本,我没有去找,不过网页版就很好用了,只需要复制粘贴进去自己想分析的数据,按照一定的格式即可,比如:
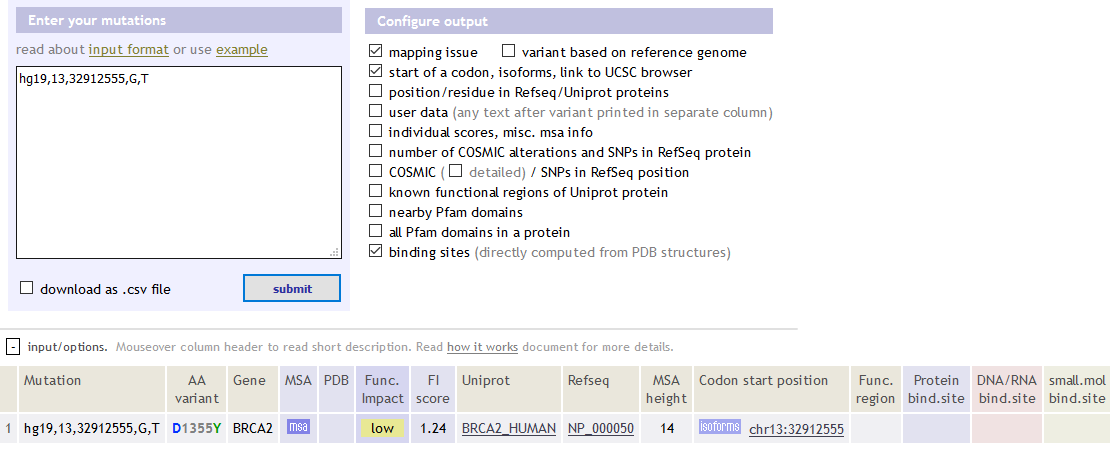
该软件就能分析出该突变位点发生在BRCA2这个基因上面,对氨基酸的改变也能写出来,对蛋白功能的改变等选项都是可以自由定制化的。
输入数据非常广泛:
The server accepts list of variants, one variant per line, plus optional text describing your variants,
in genomic coordinates, "+" strand assumed :
<genome build>,<chromosome>,<position>,<reference allele>,<substituted allele>
Genome build is optional (build 18 assumed), accepted values: 'hg18' and 'hg19'
Examples:
in genomic coordinates, "+" strand assumed :
<genome build>,<chromosome>,<position>,<reference allele>,<substituted allele>
Genome build is optional (build 18 assumed), accepted values: 'hg18' and 'hg19'
Examples:
hg19,13,32912555,G,T BRCA2
hg18,7,55178574,G,A GBM
7,55178574,G,A GBM
or in protein space: <protein ID> <variant> <text>, where <protein ID> can be :
1. Uniprot protein accession (i.e. EGFR_HUMAN)
2. NCBI Refseq protein ID (i.e. NP_005219)
examples:
EGFR_HUMAN R521K
EGFR_HUMAN R98Q Polymorphism
EGFR_HUMAN G719D disease
NP_000537 G356A
NP_000537 G360A dbSNP:rs35993958
NP_000537 S46A Abolishes phosphorylation
ID types can be mixed in one list in any way.