roadmap的官网是:http://www.roadmapepigenomics.org/
精选的129个细胞系,细胞系的介绍如下:http://www.broadinstitute.org/~anshul/projects/roadmap/metadata/EID_metadata.tab
对每个细胞系,都至少处理了5个核心组蛋白修饰数据,还有其它若干转录因子数据。
官网介绍的很详细,我就不翻译了:
The NIH Roadmap Epigenomics Mapping Consortium was launched with the goal of producing a public resource of human epigenomic data to catalyze basic biology and disease-oriented research. The Consortium leverages experimental pipelines built around next-generation sequencing technologies to map DNA methylation, histone modifications, chromatin accessibility and small RNA transcripts in stem cells and primary ex vivo tissues selected to represent the normal counterparts of tissues and organ systems frequently involved in human disease. The Consortium expects to deliver a collection of normal epigenomes that will provide a framework or reference for comparison and integration within a broad array of future studies. The Consortium also aims to close the gap between data generation and its public dissemination by rapid release of raw sequence data, profiles of epigenomics features and higher-level integrated maps to the scientific community. The Consortium is also committed to the development, standardization and dissemination of protocols, reagents and analytical tools to enable the research community to utilize, integrate and expand upon this body of data.
首先是这个网站:
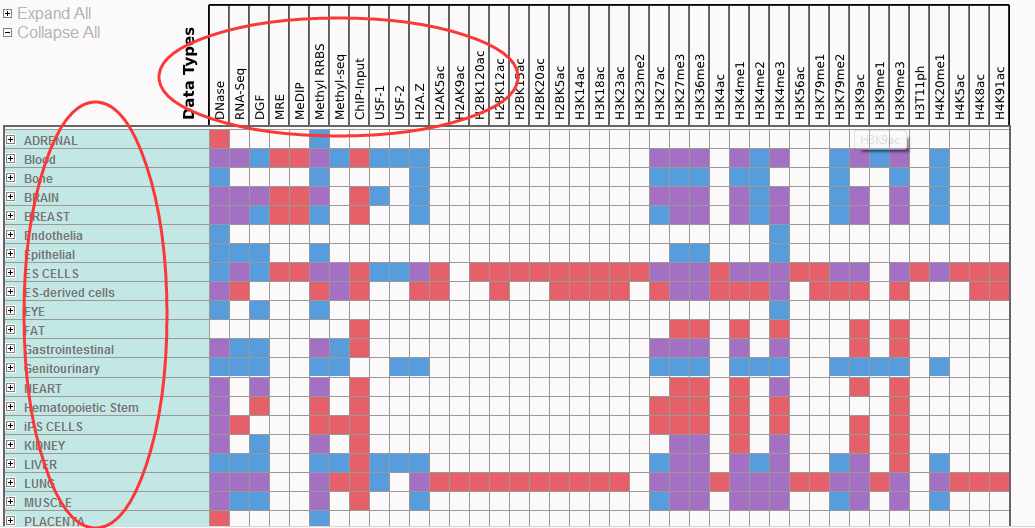
矩阵很容易看懂roadmap处理了哪些细胞系,进行了什么样的处理,数据可以直接下载。
然后我比较首先推崇broad研究所的下载方式
里面还列出了他们用过的peaks caller 工具:
http://www.broadinstitute.org/~anshul/projects/encode/preprocessing/peakcalling/ 可以看到,主要有MACS,peakranger,quest,sicer,peakseq,hotspot等等
直接进入broad分析好的peaks结果:
![[DIR]](file:///C:/Users/Jimmy/AppData/Local/YNote/data/jmzeng1314@163.com/f1dd258f825842a097d1af21461d8dc1/back.gif) |
Parent Directory | - | ||
![[DIR]](file:///C:/Users/Jimmy/AppData/Local/YNote/data/jmzeng1314@163.com/8f7a421348a94e34b4aeb53f9152b39a/folder.gif) |
broadPeak/ | 08-Feb-2015 21:00 | - | |
![[DIR]](file:///C:/Users/Jimmy/AppData/Local/YNote/data/jmzeng1314@163.com/8f7a421348a94e34b4aeb53f9152b39a/folder.gif) |
gappedPeak/ | 08-Feb-2015 21:00 | - | |
![[DIR]](file:///C:/Users/Jimmy/AppData/Local/YNote/data/jmzeng1314@163.com/8f7a421348a94e34b4aeb53f9152b39a/folder.gif) |
lowq/ | 31-Aug-2014 20:42 | - | |
![[DIR]](file:///C:/Users/Jimmy/AppData/Local/YNote/data/jmzeng1314@163.com/8f7a421348a94e34b4aeb53f9152b39a/folder.gif) |
narrowPeak/ | 08-Feb-2015 20:59 | - |
这里面有3种peaks,我现在还没有搞懂是什么意思。
接着是 iHEC存放的数据:
我还是第一次看到这个数据接口,也是以文件夹文件的形式直接浏览,根据自己的需求下载即可:
除了ENCODE计划的数据,还有Blueprint计划和roadmap计划的数据都可以下载。
| NIH Roadmap | 2014-05-29 | Click here for policies |
最后可以从圣路易斯华盛顿大学里面下载
圣路易斯华盛顿大学Washington University in St. Louis,简称(Wash U,WU)以美国国父乔治·华盛顿命名,始建于1853年2月22日,位于美国密苏里州圣路易斯市,是美国历史上建校最早也是最负盛名的“华盛顿大学”,该校在美国新闻和世界报道(US News & World Report)2014大学综合排名中名列14位。
里面有一个非常详细的页面来介绍roadmap的各种数据:http://egg2.wustl.edu/roadmap/web_portal/processed_data.html
如果你已经了解了roadmap计划,就很容易找到自己的数据,从而直接浏览器或者wget下载即可。
首先是序列比对结果下载。
onsolidated Epigenomes:36 bp mappability filtered, pooled and subsampled read alignment files:
http://egg2.wustl.edu/roadmap/data/byFileType/alignments/consolidated/
http://egg2.wustl.edu/roadmap/data/byFileType/alignments/consolidated/
Unconsolidated Epigenomes (Uniform mappability): 36 bp mappability filtered primary alignment files:
http://egg2.wustl.edu/roadmap/data/byFileType/alignments/unconsolidated/
http://egg2.wustl.edu/roadmap/data/byFileType/alignments/unconsolidated/
包括各种peaks记录文件下载
- Narrow contiguous regions of enrichment (peaks) for histone ChIP-seq and DNase-seq
- Broad domains on enrichment for histone ChIP-seq and DNase-seq)
- Data format: BroadPeak
http://egg2.wustl.edu/roadmap/data/byFileType/peaks/consolidated/broadPeak/
- Data format: GappedPeak (subset of domains containing at least one narrow peaks)
http://egg2.wustl.edu/roadmap/data/byFileType/peaks/consolidated/gappedPeak/