表达芯片大家最熟悉的当然是affymetrix系列芯片啦,而且分析套路很简单,直接用R的affy包,就可以把cel文件经过RMA或者MAS5方法得到表达矩阵。illumina出厂的芯片略微有点不一样,它的原始数据有3个层级,一般拿到的是Processed data (示例), 当仍然需要一系列的统计学方法才能提取到表达矩阵。接下来我们首先讲一讲illumina的bead 系列表达芯片基础知识吧:
illumina是大厂家,所以芯片包括人类的,小鼠以及大鼠的,然后对于人来说,经历了V1~V4的进化过程,最新版是 V4。
GEO里面是这样介绍illumina bead V4这个芯片的:
| The HumanHT-12 v4 Expression BeadChip provides high throughput processing of 12 samples per BeadChip without the need for expensive, specialized automation. The BeadChip is designed to support flexible usage across a wide-spectrum of experiments. |
可以在官网下载芯片探针的详情,manifest数据文件:http://support.illumina.com/array/array_kits/humanht-12_v4_expression_beadchip_kit/downloads.html 这些文件写清楚了芯片用的探针的详情,包括使用了哪些control探针,主要是给它自己的BeadStudio 软件来使用的。
NCBI的GEO也提供大批量的公共数据:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL10558
芯片厂家illumina本身提供数据处理软件BeadStudio, GenomeStudio,在R/bioconductor上面也有开源的包做同样的事情,Illuminaio ,beadarray,lumi
数据的前期处理有3个层次,都在bioconductor有对应的包可以来处理
Data can be in raw form, where pixel-level data are available from TIFF images, allowing the complete data processing pipeline, including image analysis, to be carried out in R. 这种图片格式的数据,基本上没有人愿意去开始处理的,TIFF格式的图片压缩包,在BeadArrayUseCases包里面附带有一个测试数据
The next level, referred to as bead-level, refers to the availability of intensity and location information for individual beads. In this format, a given probe will have a variable number of replicate intensities per sample. Processed data, where replicate intensities have been summarized and outliers removed to give a mean, a measure of variability, and a number of observations per probe in each sample, is the most commonly available format.
数据处理流程如下:
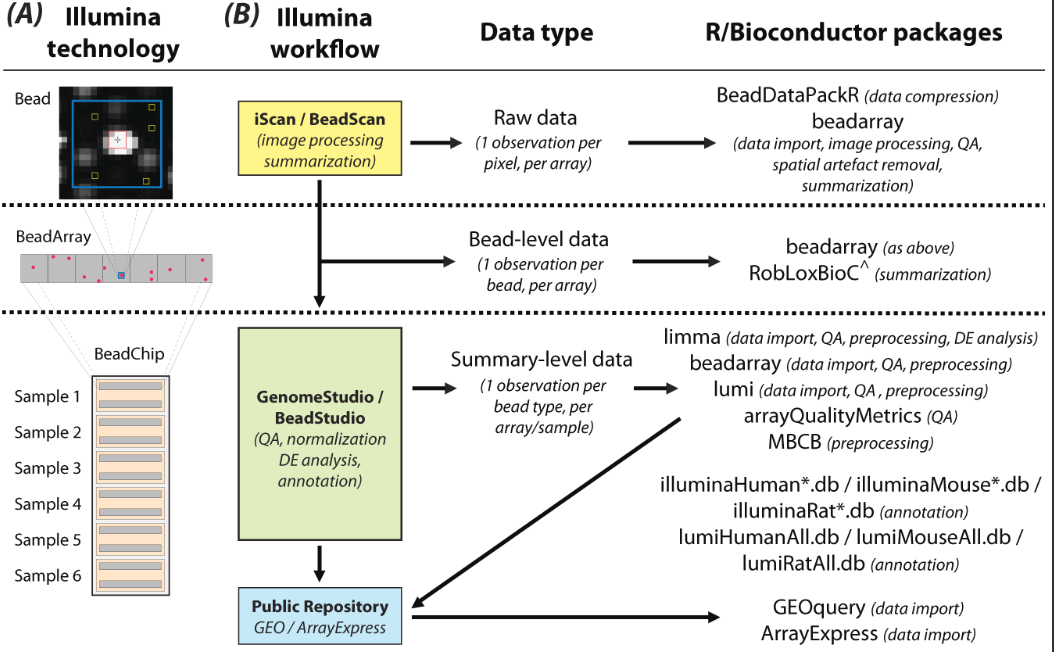
其实对芯片数据处理最重要的过程,就是如何做QC以及拿到表达量矩阵,后面的差异分析,功能富集分析其实是大同小异的。我比较喜欢用bioconductor包,会讲如何用 lumi包来处理这个芯片数据。
用bioconductor系列包来处理是最方便的,看这个教程就够了:https://bioconductor.org/packages/release/data/experiment/vignettes/BeadArrayUseCases/inst/doc/BeadArrayUseCases.pdf
数据处理流程还在plos one杂志上面发表过文章:http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1002276
BMC也有一篇:https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4486126/ 他们团队做了一个网页版工具,直接可以上传illumina芯片的原始数据去做 全套分析:http://www.arrayanalysis.org/
在R/bioconductor里面,跟人类相关的illumina beadseed芯片注释包如下:
| illuminaHumanv1.db | Mark Dunning | Illumina HumanWG6v1 annotation data (chip illuminaHumanv1) |
| illuminaHumanv2.db | Mark Dunning | Illumina HumanWG6v2 annotation data (chip illuminaHumanv2) |
| illuminaHumanv2BeadID.db | Mark Dunning | Illumina HumanWGv2 annotation data (chip illuminaHumanv2BeadID) |
| illuminaHumanv3.db | Mark Dunning | Illumina HumanHT12v3 annotation data (chip illuminaHumanv3) |
| illuminaHumanv4.db | Mark Dunning | Illumina HumanHT12v4 annotation data (chip illuminaHumanv4) |
详情可以去bioconductor官网搜索:http://www.bioconductor.org/packages/release/BiocViews.html#___AnnotationData
芯片包装如下:
