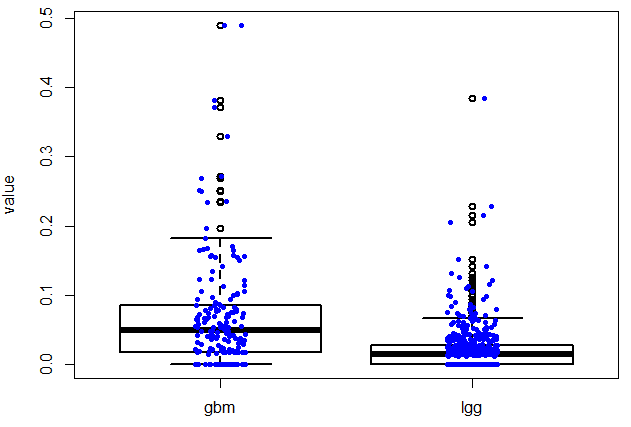
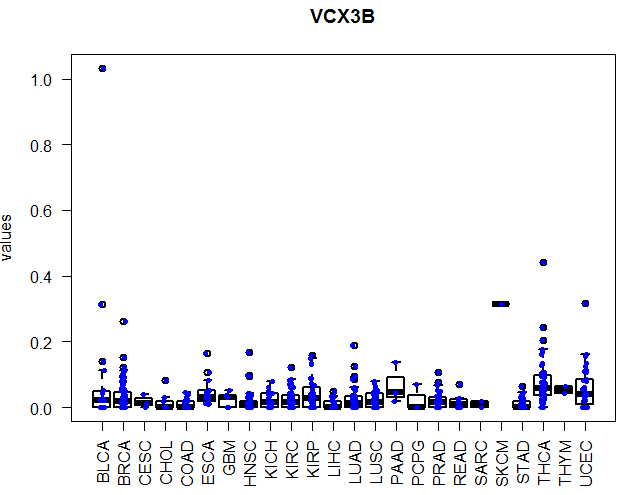
好像文章题目没有长度限制,太好了!本讲所实现的目标非常简单,如题,指定基因在不同癌种里面画boxplot,或者在所有的normal组织里面看表达量!下面是一个具体的例子:
代码如下:
稍微懂一点R的小伙伴都看得出来,只需要手动修改指定的基因,然后指定的癌症种类,就可以来容易画上面的图了,但要完成这一步,必须把前面的那一步导入mysql数据库搞懂。
TCGA表达数据的多项应用之1–下载数据并且导入mysql
rm(list=ls())searchGene = 'VCX3B';searchTable1='tumor_gbm_rpkm';searchTable2='tumor_lgg_rpkm';library(RMySQL)con <- dbConnect(MySQL(), host="127.0.0.1", port=3306, user="root", password="11111111")dbSendQuery(con, "USE gse62944")dbListTables(con)query = paste0(' select * from ', searchTable1 ,' where genesymbol = ',shQuote(searchGene)) ;gbm=dbGetQuery(con,query)query = paste0(' select * from ', searchTable2 ,' where genesymbol = ',shQuote(searchGene)) ;lgg=dbGetQuery(con,query)gbm=as.numeric(gbm[,-1]);gbm=data.frame(value=gbm,type='gbm')lgg=as.numeric(lgg[,-1]);lgg=data.frame(value=lgg,type='lgg')dat1= rbind(gbm,lgg)boxplot( value ~ type, data = dat1, lwd = 2, ylab = 'value')stripchart(value ~ type, vertical = TRUE, data = dat1,method = "jitter", add = TRUE, pch = 20, col = 'blue')
还有很多其它的应用,重点就是如何从sql里面提取数据并可视化而已
比如上面那个在正常表达量矩阵里面查询,多种癌旁组织合并起来画图!
sqlTable = 'normalrpkm';sqlQuery=paste0(' select * from ', sqlTable ,' where genesymbol = ',shQuote(searchGene))normalExpression=dbGetQuery(con,sqlQuery)normalExpression= normalExpression[,-length(normalExpression)]normalExpression = data.frame(sampleID=names(normalExpression),values=as.numeric(normalExpression))normalCancerType2amples=dbGetQuery(con,'select * from normalcancertype2amples')normalCancerType2amples$sampleID=gsub("-",".", normalCancerType2amples$sampleID)dat2 = merge(normalExpression,normalCancerType2amples,by='sampleID')boxplot( values ~ CancerType, data = dat2, lwd = 2, ylab = 'values',las=2,main=searchGene)stripchart(values ~ CancerType, vertical = TRUE, data = dat2,method = "jitter", add = TRUE, pch = 20, col = 'blue')