TET2突变是如何引起超甲基化
癌症病人体内会检测到不正常的甲基化现象。
TET2可以氧化5mC成为5hmC,进而通过其它机制形成5fC和5caC。
很多血液肿瘤病人的TET2基因突变了,同时会显示出全局的5hmC水平下降。
有趣的是,全局的5hmC水平下降同样发生在很多实体肿瘤病人身上,但是那些病人很少有TET2突变发生。
那么,TET2突变,或者全局的5hmC水平下降,是如何导致启动子区域的CG岛的甲基化水平上升的呢?
有其它文献报道 hypermethylation和oxidative stress (OS)有关系
作者认为 oxidative stress (OS) 在其中起了关键的作用。
关键结论
- We now demonstrate TET2 forms ‘‘Yin-Yang’’ complexes with DNMTs and is targeted to chromatin during OS.
- TET2 actively removes abnormal DNAm induced by OS in promoter CGIs as well as enhancers by converting unwanted 5mC to 5hmC.
- Long-term reduction of TET2 caused even more DNA hypermethylation on these gene promoters and enhancers.
数据
作者用了
Agilent-026652 Whole Human Genome Microarray 4x44K v2和Illumina HumanMethylation450 BeadChip来联合表达量和甲基化进行分析。 - GSE81426
Genome wide DNA methylation profiling of A2780 cells -
untreated (mock)
- treated with H2O2 for 30 min (H2O2 30 min)
- treated with H2O2 for 30 min with additional 2.5 hour resting ( H2O2 3h) .
-
GSE81427
Genome wide DNA methylation profiling of A2780 cells - 1) infected with control virus, no H2O2 (Scr_mock),
- 2) infected with control virus with H2O2 treatment (30 min plus 2.5 h resting) (Scr_H2O2),
- 3) infected with shTET2 virus, no H2O2 (shTET2_mock)
- _4) infected with shTET2 virus, with H2O2 treatment (30 min plus 2.5 h resting) (shTET2_H2O2, two biological replicates).
- GSE79808
- GSE83750
- GSE83751
数据分析要点及结论
主要是集中在 Figure 7. TET2 Protects against Abnormal DNAm
图1
把TET2基因使用LentiCRISPR病毒KO掉前后的A2780细胞系,选取那些表达量下调非常显著的基因集。然后再根据它们这些基因的甲基化芯片的promoter CGI 探针的 β值来分成3类:DNAm of unmethylated (b < 0.25) and intermediately methylated (0.25 < b < 0.75) ,分析发现它们的甲基化程度显著上升。
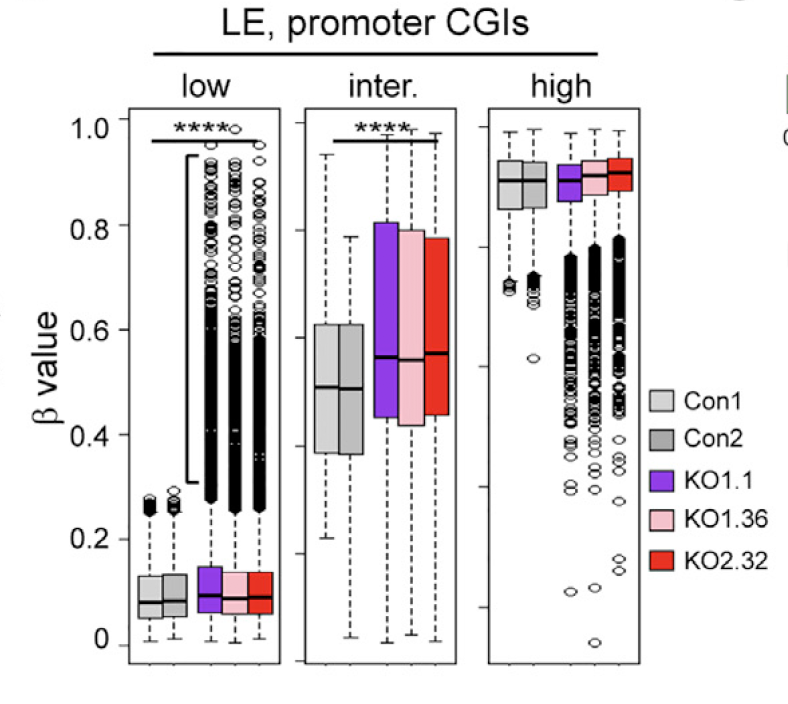
图2
还是针对那些TET2基因敲除后表达下降的基因,热图展现它们的甲基化芯片的promoter CGI 探针的 β值,共2947个探针。
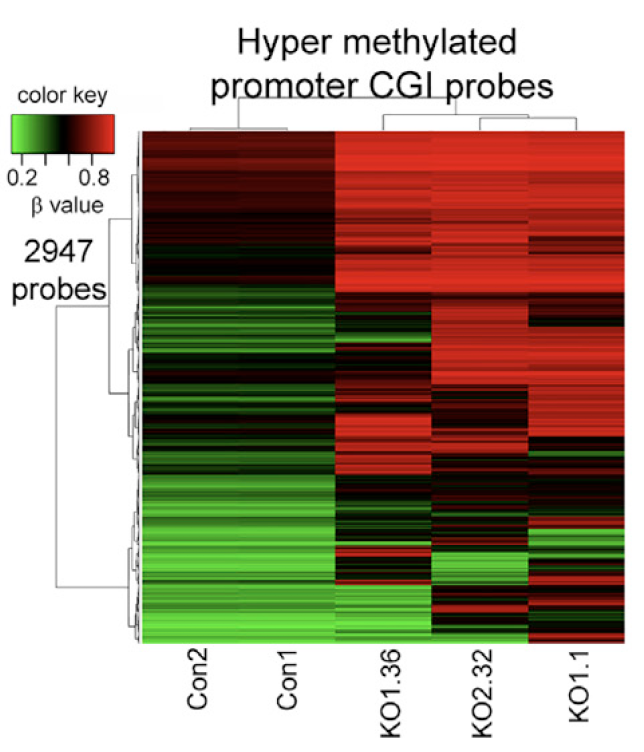
图3
把TET2基因敲除后那些超甲基化的1406个基因跟以前发表的bivalent genes in hESCs, and cancer specific hypermethylated genes.基因集用韦恩图展现交叉情况。
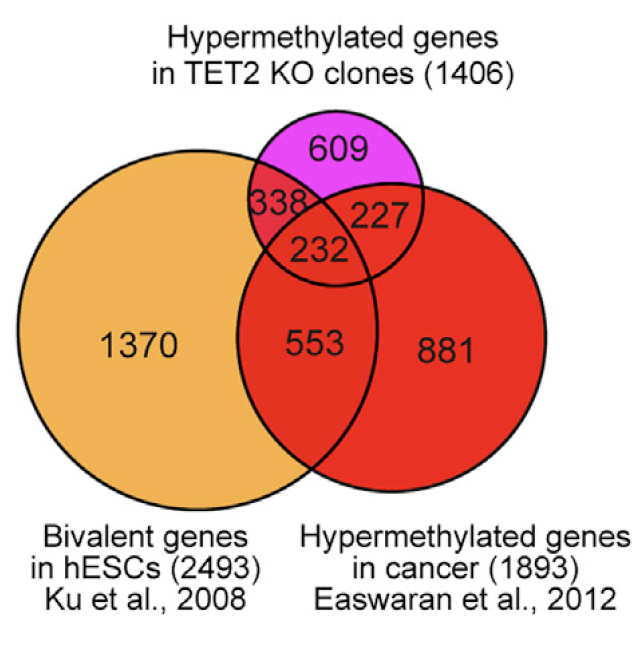
图4
把TET2基因敲除后那些超甲基化的1406个基因进行GO富集分析
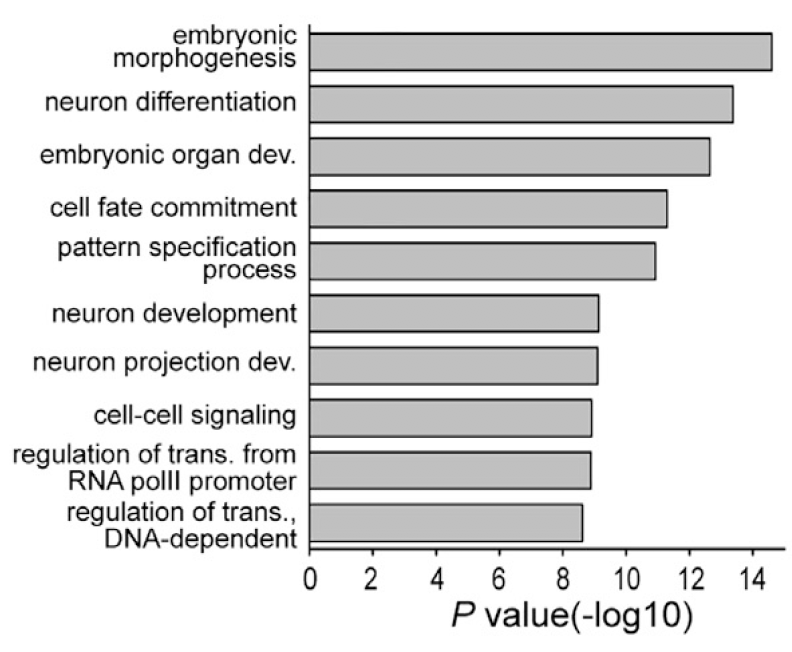
图5
把TET2基因敲除前后细胞表达量的变化以及它们对应的promoter CGI 探针的 β值变化的散点图
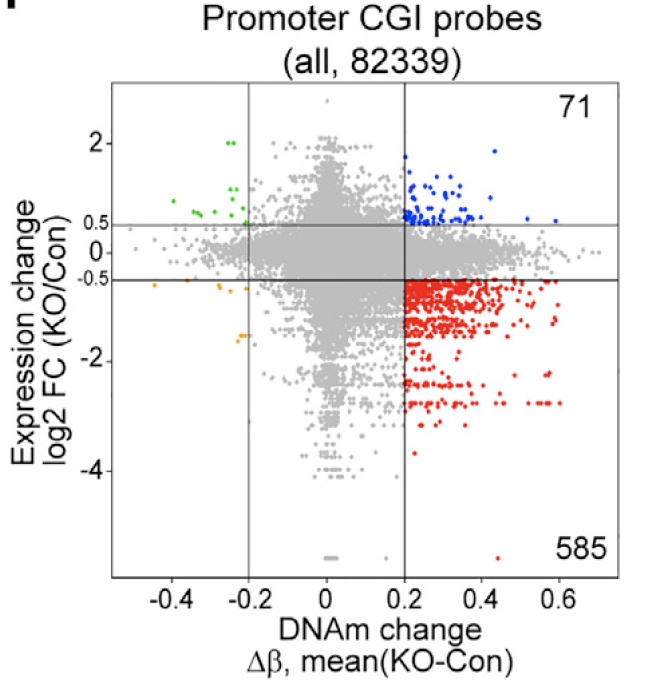
同时也分析了增强子区域:
2,107 hypermethylated enhancer regions identified by Rasmussen et al. (2015) to similar subgroups based on their basal methylation levels and found a very similar pattern for gains of DNAm in their TET2 KO scenario