单细胞转录组探索CRC病人的一致性
文章是:Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 2017 May;49(5):708-718. PMID: 28319088
单细胞转录组,使用的是GPL11154Illumina HiSeq 2000 (Homo sapiens) ,数据都上传到了:GSE81861
| BioProject | PRJNA323703 |
|---|---|
| SRA | ERP016958 |
既有病人的单细胞转录组数据,同时也有细胞系的数据做验证。
- 1,591 single cells from 11 colorectal cancer patients,包括 969肿瘤部位细胞以及 622 癌旁细胞。严格过滤后只剩下:375 tumor cells and 215 normal mucosa cells
- 630 single cells from 7 cell lines,过滤后剩下561个
- 83 A549 cells,
- 65 H1437 cells,
- 55 HCT116 cells,
- 23 IMR90 cells,
- 96 K562 cells,
- 134 GM12878 cells (38 from batch 1, 96 from batch 2)
- 174 H1 cells (96 from batch 1, 78 from batch 2).
上游测序数据没有必要重新下载分析了,可以直接使用作者上传的表达矩阵:
| Supplementary file | Size | Download | File type/resource |
|---|---|---|---|
| GSE81861_CRC_NM_all_cells_COUNT.csv.gz | 3.2 Mb | (ftp)(http) | CSV |
| GSE81861_CRC_NM_all_cells_FPKM.csv.gz | 4.7 Mb | (ftp)(http) | CSV |
| GSE81861_CRC_NM_epithelial_cells_COUNT.csv.gz | 2.5 Mb | (ftp)(http) | CSV |
| GSE81861_CRC_NM_epithelial_cells_FPKM.csv.gz | 4.0 Mb | (ftp)(http) | CSV |
| GSE81861_CRC_tumor_all_cells_COUNT.csv.gz | 4.3 Mb | (ftp)(http) | CSV |
| GSE81861_CRC_tumor_all_cells_FPKM.csv.gz | 7.9 Mb | (ftp)(http) | CSV |
| GSE81861_CRC_tumor_epithelial_cells_COUNT.csv.gz | 3.6 Mb | (ftp)(http) | CSV |
| GSE81861_CRC_tumor_epithelial_cells_FPKM.csv.gz | 6.5 Mb | (ftp)(http) | CSV |
| GSE81861_Cell_Line_COUNT.csv.gz | 13.1 Mb | (ftp)(http) | CSV |
| GSE81861_Cell_Line_FPKM.csv.gz | 28.9 Mb | (ftp)(http) | CSV |
| GSE81861_GEO_EGA_ID_match.csv.gz | 14.4 Kb | (ftp)(http) | CSV |
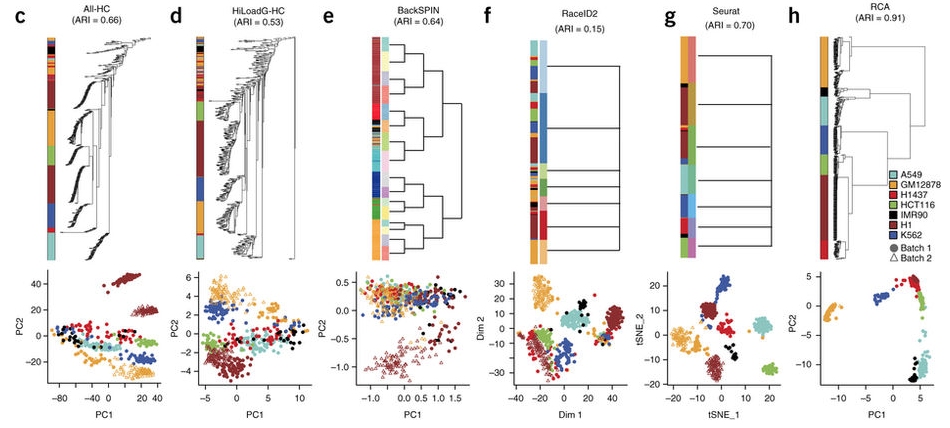
作者认为全文最重要的是开发了一个挖掘细胞类型的算法:reference component analysis (RCA) 优于其它现有的算法。可以把cancer-associated fibroblasts (CAFs)继续分成两个类别。对比的算法包括:
- hierarchical clustering using all expressed genes (All-HC)
- hierarchical clustering using principal-component analysis (PCA)-based feature selection (HiLoadG-HC)
- BackSPIN
- RaceID2
- Seurat
- three additional methods based on selection of genes with highly variable expression (VarG-HC, VarG-PCAproj-HC and VarG-tSNEproj-HC).
使用 adjusted Rand index (ARI) 指标来评价各个聚类算法的优劣。结果发现自己开发的RCA表现超常!!!
 然了,还在 Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA–seq. Science 352, 189–196 (2016). 文章的数据里面做了验证。
然了,还在 Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA–seq. Science 352, 189–196 (2016). 文章的数据里面做了验证。
背景知识
肿瘤异质性很重要,单细胞转录组测序很厉害,以前的研究根据单细胞转录组表达矩阵进行分类的算法不够好,所以他们开发reference component analysis (RCA) , 而且 Colorectal cancer (CRC) 疾病非常严重,需要探索。
根据细胞系单细胞表达数据探索算法
630个细胞的表达数据,过滤后剩下561个,这里使用Fragments per kilobase per million reads (FPKM)来进行表达定量。因为其上游处理走的是TOPHAT2+CUFFLINKS流程。
单细胞过滤策略
rate of exonic reads (ROER) 需要大于5%
number of detected genes (NODG) 需要大于1000, 基因的FPKM ≥1才能算被检测到了。
Exonic reads (ER) 要大于0.1Million
管家基因: TFRC, ACTB, RPLP0, PGK1, GAPDH, LDHA, NONO, B2M, GUSB and PPIH.
RCA算法细节
首先从 BioGPS数据库里面下载两个数据集: HumanU133A/GNF1H Gene Atlas and the Primary Cell Atlas ,从中挑选 A total of 4,717 genes were selected as features for GNF1H and 5,209 genes were selected for the Primary Cell Atlas. 还使用了 WGCNA 算法。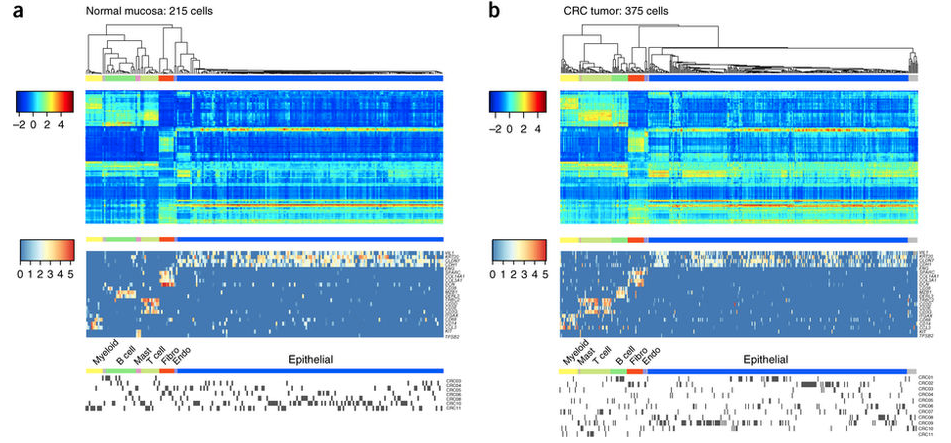
还使用了一些其它公共数据:TCGA, GSE14333, the PRECOG database, and GSE33113, GSE37892 and GSE39582 来验证单细胞转录组得到的基因集(The 'fibroblast-like' signature )是否能显著的区分CRC病人的生存情况。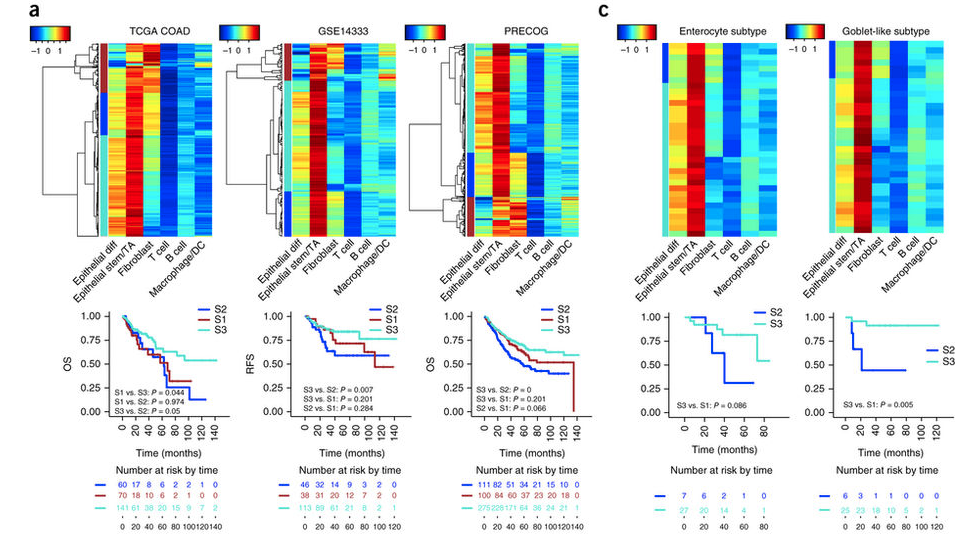
需要了解一些细胞类型的 known markers
- epithelial cells (VIL1, KRT20, CLDN7, CDH1)
- endothelial cells (Endo; ENG)
- fibroblasts (Fibro; SPARC, COL14A, COL3A1, DCN)
- B cells (CD38, MZB1, DERL3)
- T cells (TRBC2, CD3D, CD3E, CD3G)
- myeloid cells (ITGAX, CD68, CD14, CCL3)
- mast cells (KIT, TPSB2)
做成了一个R包供使用:RCA R package, https://github.com/GIS-SP-Group/RCA.