有成熟的R包可以把bam文件读入R,比如Rsamtools,很简单的代码:
library(Rsamtools)
bamFile="alignResults.BAM"
quickBamFlagSummary(bamFile)
# https://kasperdanielhansen.github.io/genbioconductor/html/Rsamtools.html
bam <- scanBam(bamFile)
bam
但是把读入的数据变成grange对象就需要一点点技巧:
names(bam[[1]])
tmp=as.data.frame(do.call(cbind,lapply(bam[[1]], as.character)))
tmp=tmp[tmp$flag!=4,] # 60885 probes
# intersect() on two GRanges objects.
library(GenomicRanges)
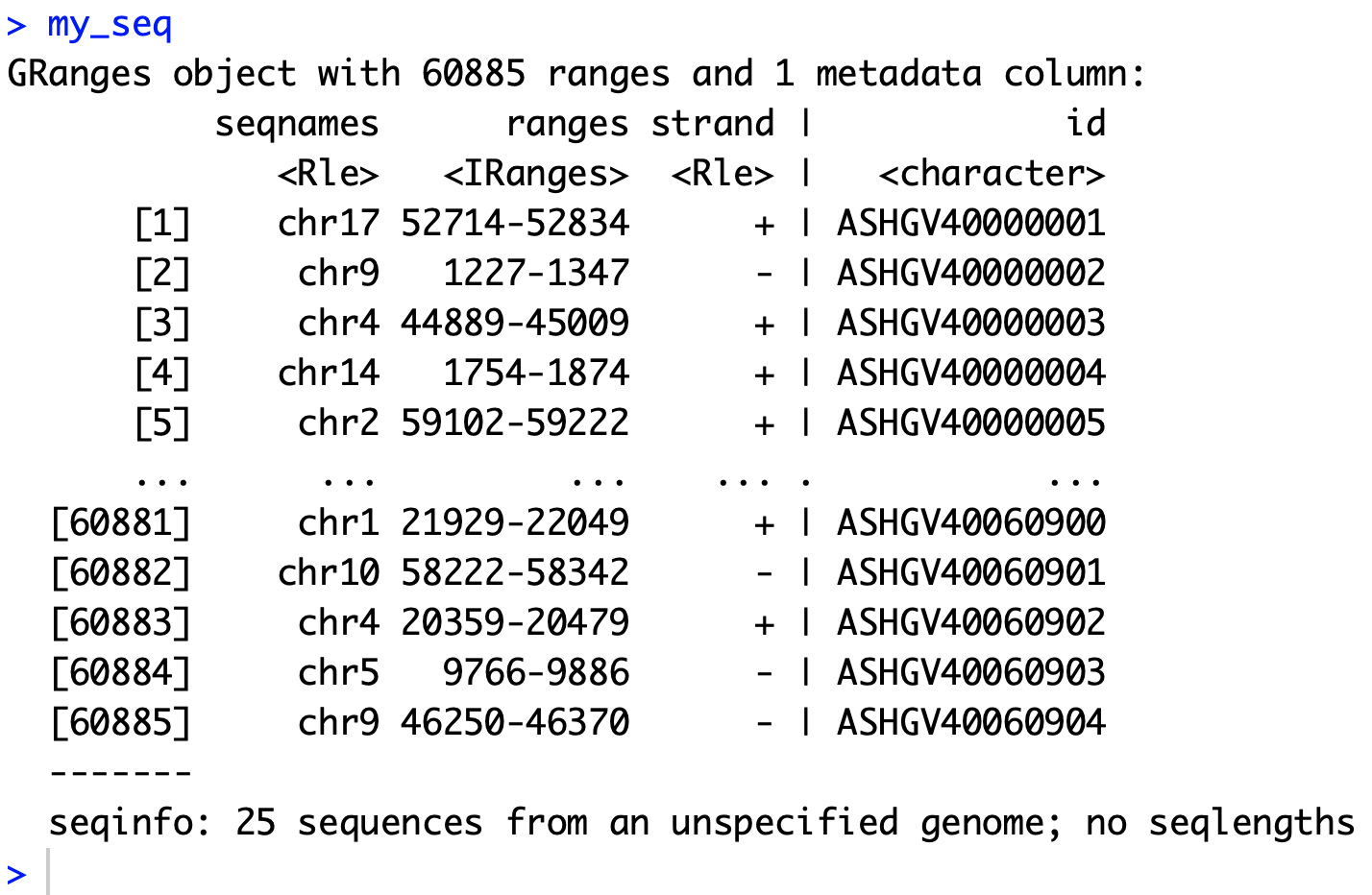
my_seq <- with(tmp, GRanges(as.character(rname),
IRanges(as.numeric(pos)-60, as.numeric(pos)+60),
as.character(strand),
id = as.character(qname)))
得到对象如下: