最近在答疑群里收到一个很经典的提问,就是:
- 请问各位老师,GPL570芯片中应该有部分基因是LncRNA,能否通过基因重注释的方式把有意义的LncRNA筛选出来呢?R语言能否实现呢?
而且学生特别的好学,已经懂得去搜索我们已有的1.3万篇教程,找到了芯片探针序列重新注释的流程,但是我昨天就说到过:芯片探针序列的基因注释已经无需你自己亲自做了, 肯定是学员没有追我们的公众号最新教程,不过这个不能怪他。这个是公众号的弊端,太多冗余信息让大家分心,与我们真正的知识分享初衷背道而驰了。
所以呢,其实使用我们的包,安装方法说到过:芯片探针序列的基因注释已经无需你自己亲自做了, ,使用起来也非常简单:
library(AnnoProbe)
ids=idmap('GPL570',type = 'soft')
head(ids)
仅仅是一句话,就拿到了这个平台的探针的注释信息,如下:
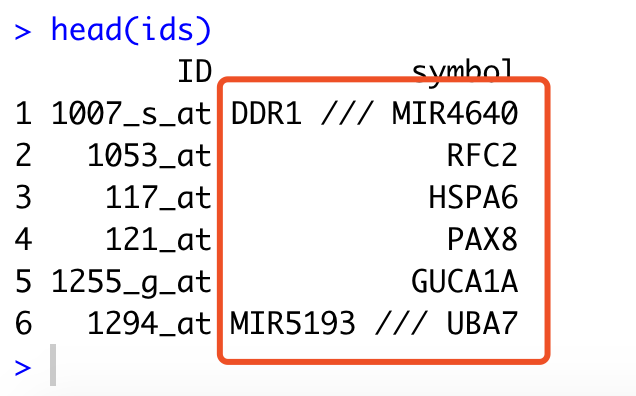
但是呢,我们还是探索一下,因为这个是下载的GPL的soft文件里面的注释信息,所以可以看到是有一些探针居然是对应多个基因,其实是因为这些基因本身坐标就是有overlap,所以呢, 探索的代码就会稍微复杂一点。
ids=ids[nchar(ids[,2])>1,]
ids1=ids[grepl('///',ids[,2]),]
ids2=ids[!grepl('///',ids[,2]),]
# 我觉得下面的函数写的很差,运行太慢
tmp = do.call(rbind,apply(ids1,1,function(x){
x[1];x[2]
data.frame(ID=x[1],symbol=strsplit(x[2],' /// ')[[1]])
})
)
ids=rbind(ids2,tmp)
anno=annoGene(ids$symbol,"SYMBOL")
ids=merge(ids,anno,by.x = 'symbol',by.y='SYMBOL',all.x = T)
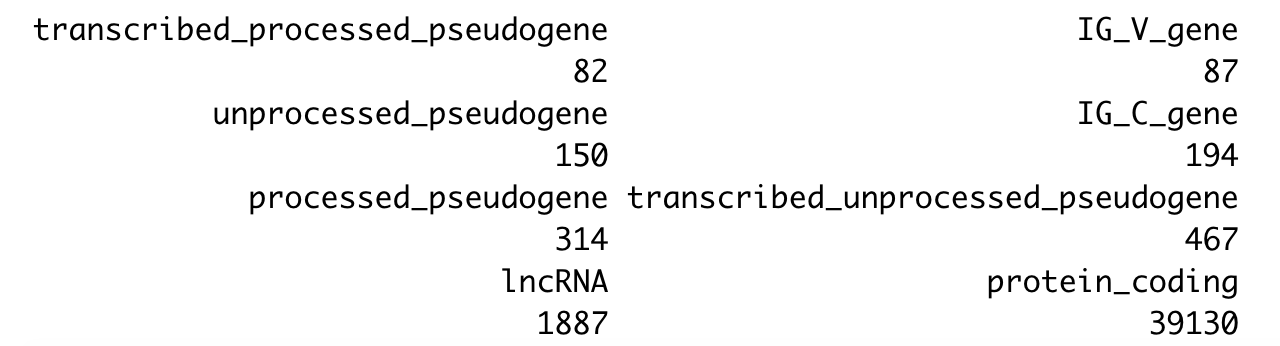
sort(table(ids$biotypes))
可以看到,五万多个探针里面,真正的蛋白编码基因的探针只有4万,剩余的一万多都是可以进行探索的。
但是呢,这个并不是最佳的选择,因为我们并没有对这个GPL平台的探针的碱基序列进行参考基因组比对后,自己重新注释,而还是使用的GPL里面的soft文件的信息。
我们看看其它芯片文献里面的GPL570探针ID的基因注释信息
比如Published: 12 March 2019的文章:Identification of Key Long Non-Coding RNAs in the Pathology of Alzheimer’s Disease and their Functions Based on Genome-Wide Associations Study, Microarray, and RNA-seq Data
Briefly, we first downloaded the reference sequences of these potentially AD-related lncRNAs in FASTA format from NONCODE database .
Second, probe sets of the microarrays were aligned to the lncRNA sequences using SeqMap tool, and the lncRNA-specific probe sets were obtained which contain at least four probes uniquely mapped to the lncRNA sequences without mismatch.
或者
Briefly, probe sets of HG-U133_Plus_2.0 array were aligned to the human genome (GRCh38) and lncRNA gene sequence from GENCODE (release 23) using SeqMap tool with no mismatch [49].
Then lncRNA-specific probes were obtained by mapping the genomic locations of probes to the genomic locations of lncRNAs.
Finally, expression data of 2332 lncRNA were obtained for further analysis.
又或者
we obtained 3215 probes (probe sets) covering 2330 lncRNAs for Affymetrix HG-U133_Plus_2.0 array and 855 probes (probe sets) covering 663 lncRNAs for Affymetrix HG-U133A array, respectively. The expression data of multiple probes (probe sets) mapping to the same lncRNA were integrated by using the arithmetic mean to represent the expression level of single lncRNA.
total of 598 probes corresponding to 452 lncRNAs were obtained for the HG-U133A microarray, while 5,654 probes were matching with 3,793 lncRNAs in the HG-U133 Plus 2.0 microarray.
又或者
Briefly, the probe sets of Affymetrix HG‐U133 Plus 2.0 were retrieved from the Affymetrix website (http://www.affymetrix.com). We then re‐mapped those probes to the chromosomal positions of the ncRNAs derived from GENCODE (release 24, GRCh38) with no mismatch 14. A total of 2380 probes and 2118 corresponding ncRNA genes were obtained. When multiple probes mapped to the same ncRNA, we used the arithmetic mean of the probe intensities.
参考文献:
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5341997/
- https://www.ncbi.nlm.nih.gov/pubmed/26183581
- https://www.ncbi.nlm.nih.gov/pubmed/26362431
- https://www.ncbi.nlm.nih.gov/pubmed/27105492
- https://onlinelibrary.wiley.com/doi/full/10.1002/cam4.1047
- http://dx.doi.org/10.4048/jbc.2018.21.e39
既然每个文献都不一样
而且大部分人是没办法自主注释的,所以我们理论上应该是有一个平台代替大家做全部的芯片探针的碱基序列的重新注释。
我们前面提到的:芯片探针序列的基因注释已经无需你自己亲自做了 里面的AnnoProbe包已经在帮大家一个个的注释啦。
敬请期待全部GPL的重新注释。