我们说到过,安装R包基本上是缺啥就安装啥即可,但是总有一些让你头疼的,比如我们介绍过在Ubuntu安装单细胞的3大R包,就非常精彩,视频在:https://www.bilibili.com/video/av63988858
最近有粉丝反映跟着我们的教程:使用inferCNV分析单细胞转录组中拷贝数变异 里面的R包安装总是失败,值得单独写教程强调一下这个解决方案。
报错是rjags这个包失败
> library(infercnv)
Error: package or namespace load failed for ‘infercnv’:
.onLoad failed in loadNamespace() for 'rjags', details:
call: dyn.load(file, DLLpath = DLLpath, ...)
error: unable to load shared object '/Library/Frameworks/R.framework/Versions/3.6/Resources/library/rjags/libs/rjags.so':
dlopen(/Library/Frameworks/R.framework/Versions/3.6/Resources/library/rjags/libs/rjags.so, 10): Library not loaded: /usr/local/lib/libjags.4.dylib
Referenced from: /Library/Frameworks/R.framework/Versions/3.6/Resources/library/rjags/libs/rjags.so
Reason: image not found
>
如果再强行安装rjags倒是可以成功,但是加载仍然失败
> install.packages("rjags")
trying URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.6/rjags_4-10.tgz'
Content type 'application/x-gzip' length 324806 bytes (317 KB)
==================================================
downloaded 317 KB
The downloaded binary packages are in
/var/folders/2x/l4fzfvy17gz94j1sz6klhyjh0000gn/T//RtmpCWo2jg/downloaded_packages
其实是因为你的电脑操作系统缺失这个
-
如果是macos呢,可以:brew install jags
-
如果是Windows呢,点击 https://sourceforge.net/projects/mcmc-jags/files/JAGS/4.x/Windows/JAGS-4.3.0.exe 下载安装即可
-
至于Linux,稍微复杂一点,但是我们有conda,不过既然可以conda了,还不如一步到位,直接conda你想要的软件即可。
比如我的苹果电脑,安装后
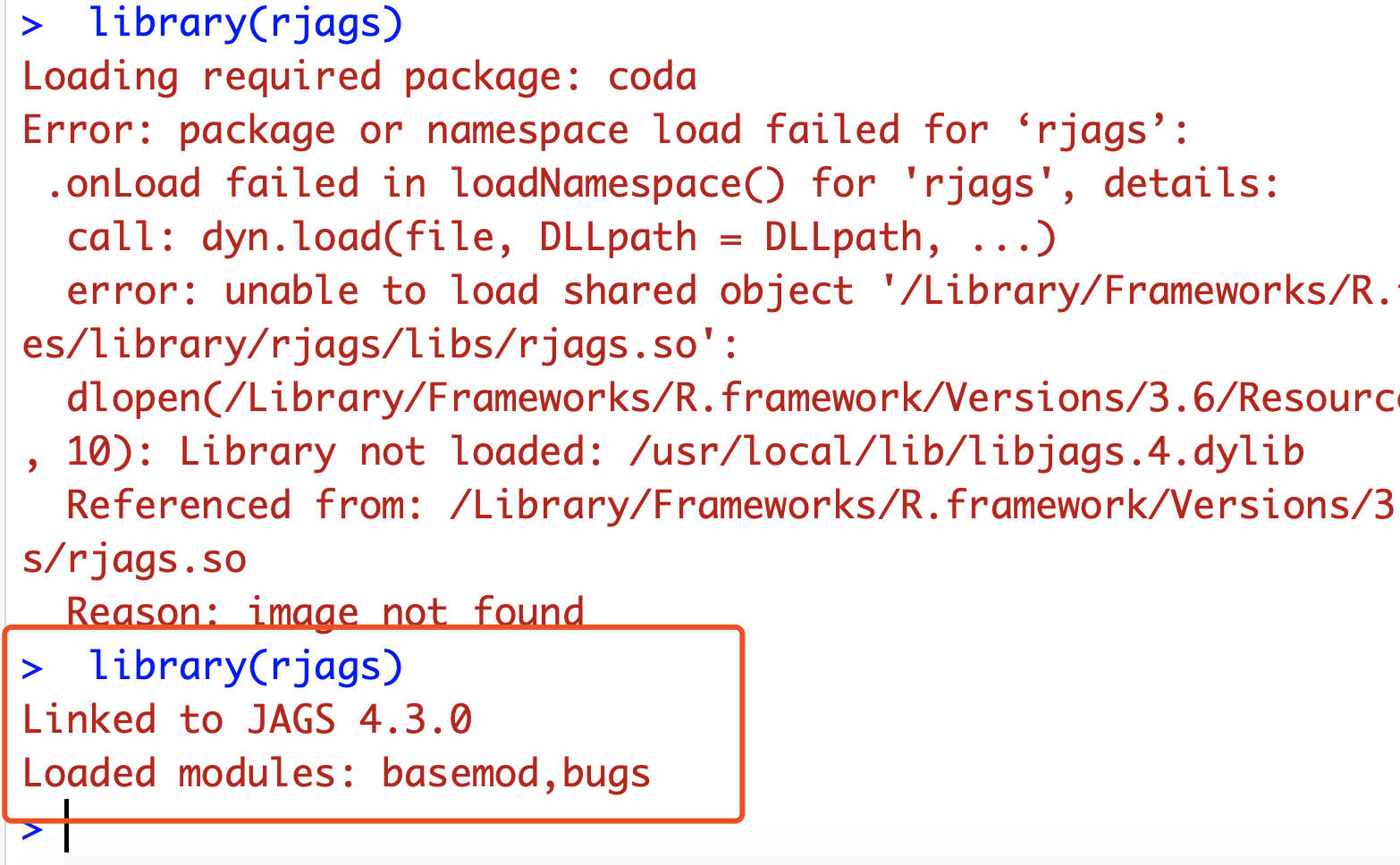
就成功加载啦。
试用一下infercnv
走一下 https://github.com/broadinstitute/inferCNV/wiki 的示例代码即可:
library(infercnv)
infercnv_obj = CreateInfercnvObject(raw_counts_matrix=system.file("extdata", "oligodendroglioma_expression_downsampled.counts.matrix.gz", package = "infercnv"),
annotations_file=system.file("extdata", "oligodendroglioma_annotations_downsampled.txt", package = "infercnv"),
delim="\t",
gene_order_file=system.file("extdata", "gencode_downsampled.EXAMPLE_ONLY_DONT_REUSE.txt", package = "infercnv"),
ref_group_names=c("Microglia/Macrophage","Oligodendrocytes (non-malignant)"))
infercnv_obj = infercnv::run(infercnv_obj,
cutoff=1, # cutoff=1 works well for Smart-seq2, and cutoff=0.1 works well for 10x Genomics
out_dir=tempfile(),
cluster_by_groups=TRUE,
denoise=TRUE,
HMM=TRUE)
因为是示例数据,数据量不大,如下所示:

这个教程的重点就是使用CreateInfercnvObject来构建一个对象,去运行infercnv::run步骤即可,大家需要注意的是输入文件的准备。把你自己的文件,按照这3个来制作即可。
输出文件也有点多,因为设置输出到临时目录,所以就不一一解读了。
## Making the final infercnv heatmap ##
INFO [2019-12-30 12:14:27] ::plot_cnv:Start
INFO [2019-12-30 12:14:27] ::plot_cnv:Current data dimensions (r,c)=8508,184 Total=1569112.28502327 Min=0.530812412297805 Max=1.98172179650671.
INFO [2019-12-30 12:14:27] ::plot_cnv:Depending on the size of the matrix this may take a moment.
INFO [2019-12-30 12:14:28] plot_cnv(): auto thresholding at: (0.649902 , 1.350098)
INFO [2019-12-30 12:14:28] plot_cnv_observation:Start
INFO [2019-12-30 12:14:28] Observation data size: Cells= 142 Genes= 8508
INFO [2019-12-30 12:14:28] clustering observations via method: ward.D
INFO [2019-12-30 12:14:28] plot_cnv_observation:Writing observation groupings/color.
INFO [2019-12-30 12:14:36] Colors for breaks: #00008B,#24249B,#4848AB,#6D6DBC,#9191CC,#B6B6DD,#DADAEE,#FFFFFF,#EEDADA,#DDB6B6,#CC9191,#BC6D6D,#AB4848,#9B2424,#8B0000
INFO [2019-12-30 12:14:36] Quantiles of plotted data range: 0.649901584270269,1.00394102388827,1.00394102388827,1.00394102388827,1.35009841572973
INFO [2019-12-30 12:14:36] plot_cnv_references:Writing observation data to /var/folders/2x/l4fzfvy17gz94j1sz6klhyjh0000gn/T//RtmpCWo2jg/file147efe28e336/infercnv.observations.txt
INFO [2019-12-30 12:14:37] plot_cnv_references:Start
INFO [2019-12-30 12:14:37] Reference data size: Cells= 42 Genes= 8508
INFO [2019-12-30 12:14:37] plot_cnv_references:Number reference groups= 2
INFO [2019-12-30 12:14:37] plot_cnv_references:Plotting heatmap.
INFO [2019-12-30 12:14:39] Colors for breaks: #00008B,#24249B,#4848AB,#6D6DBC,#9191CC,#B6B6DD,#DADAEE,#FFFFFF,#EEDADA,#DDB6B6,#CC9191,#BC6D6D,#AB4848,#9B2424,#8B0000
INFO [2019-12-30 12:14:39] Quantiles of plotted data range: 0.693386142642158,1.00394102388827,1.00394102388827,1.00394102388827,1.35009841572973
INFO [2019-12-30 12:14:39] plot_cnv_references:Writing reference data to /var/folders/2x/l4fzfvy17gz94j1sz6klhyjh0000gn/T//RtmpCWo2jg/file147efe28e336/infercnv.references.txt