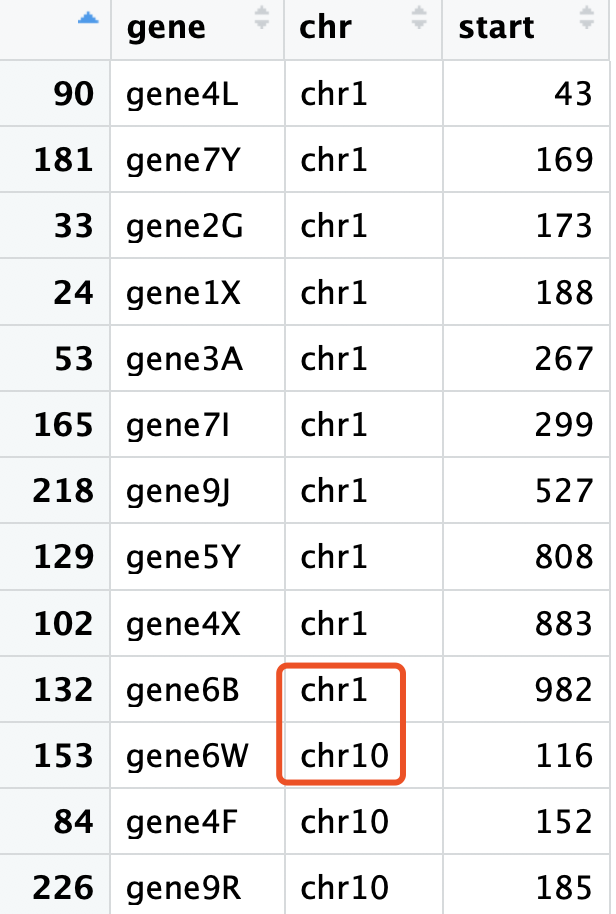
使用下面的代码模拟数据
df=do.call(rbind,lapply(1:10, function(i){
data.frame(gene=paste0('gene',i,LETTERS),
chr=sample(paste0('chr',1:22),26,replace = T),
start= sample(1:1000,26))
}))
df=df[with(df,order(chr,start)),]
df$chr=as.factor(df$chr)
plot(df$chr,df$start,las=2)
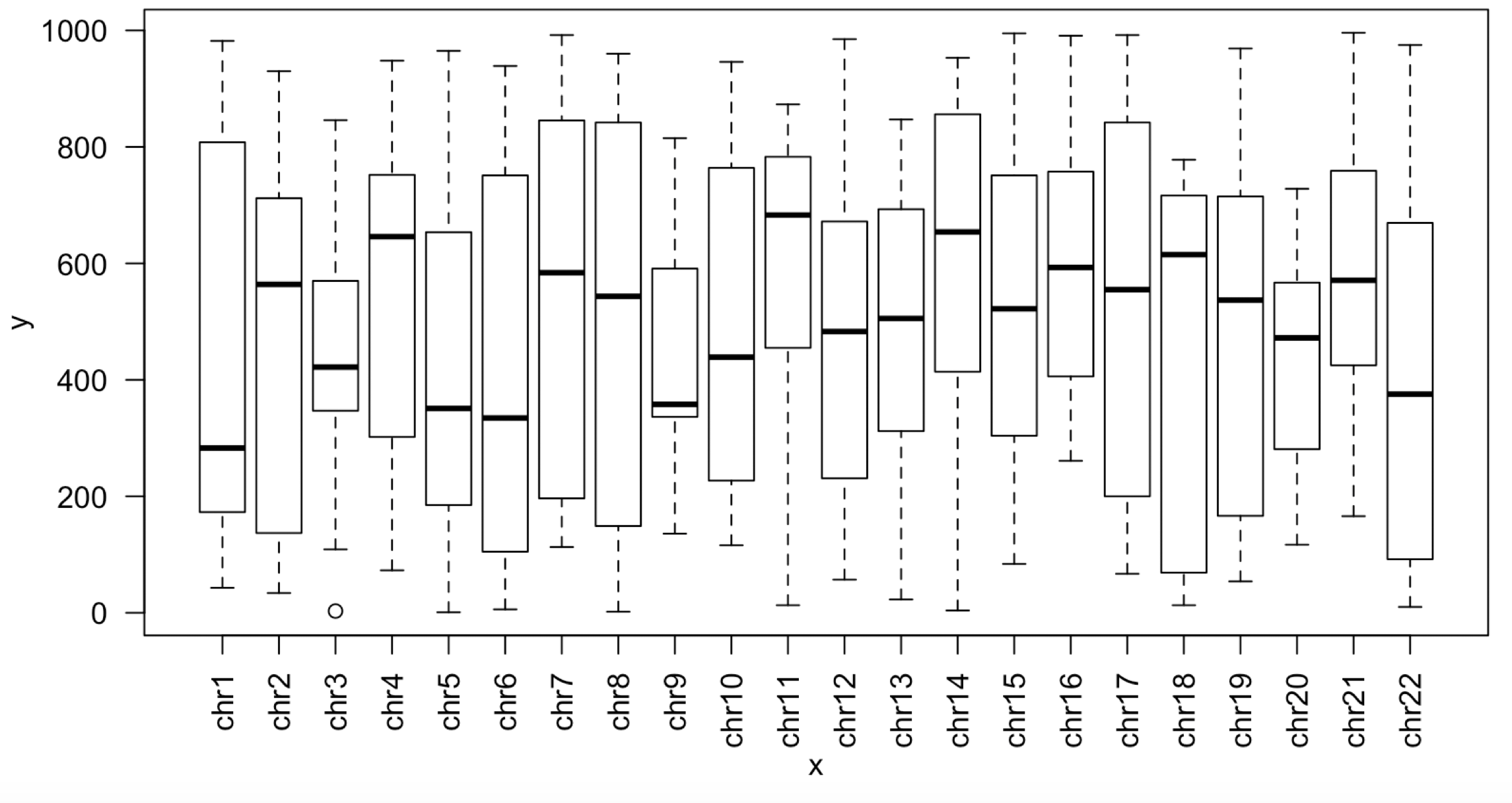
首先我们的排序并没有按照染色体顺序,而是
> levels((df$chr))
[1] "chr1" "chr10" "chr11" "chr12" "chr13" "chr14" "chr15" "chr16"
[9] "chr17" "chr18" "chr19" "chr2" "chr20" "chr21" "chr22" "chr3"
[17] "chr4" "chr5" "chr6" "chr7" "chr8" "chr9"
>
这个顺序显然不是我们想要的:
绘制出来的boxplot如下:
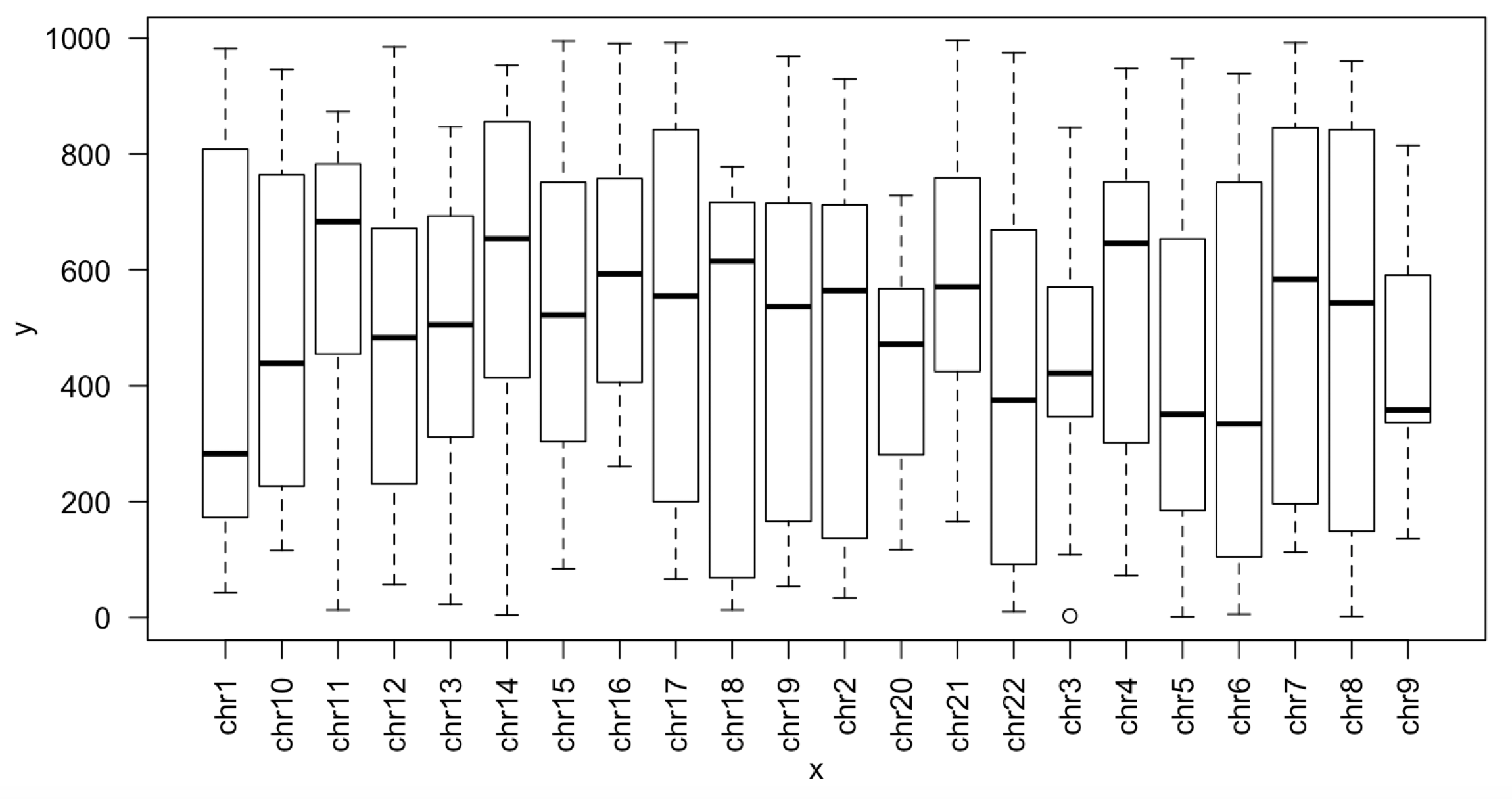
那么简单的一个转换即可,代码如下:
df$chr=factor(df$chr,paste0('chr',1:22),ordered = T)
df=df[with(df,order(chr,start)),]
plot(df$chr,df$start,las=2)
绘制出来的新的boxplot如下: