最近在整理各个癌症的单细胞转录组数据,发现 Cell. 2018 Aug 23,题目是:Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment,文章的数据量本身就很大:
- We profiled 45,000 immune cells from eight breast carcinomas, as well as matched normal breast tissue, blood, and lymph nodes, using single-cell RNA-seq.
- Analysis of paired single-cell RNA and T cell receptor (TCR) sequencing data from 27,000 additional T cells revealed the combinatorial impact of TCR utilization on phenotypic diversity.
里面清清楚楚写到作者认为cellranger虽然是10x官方软件也未必得全信它:We note that Cell Ranger, the most commonly used 10× pipeline, carries out an extreme version of this redesign: it removes any gene that is not protein coding. We believe that this is too harsh: it excludes numerous transcribed pseudogenes and lincRNA which have been previously shown to be expressed, have biological functionality, and be detectable in scRNA-seq.
如下图:
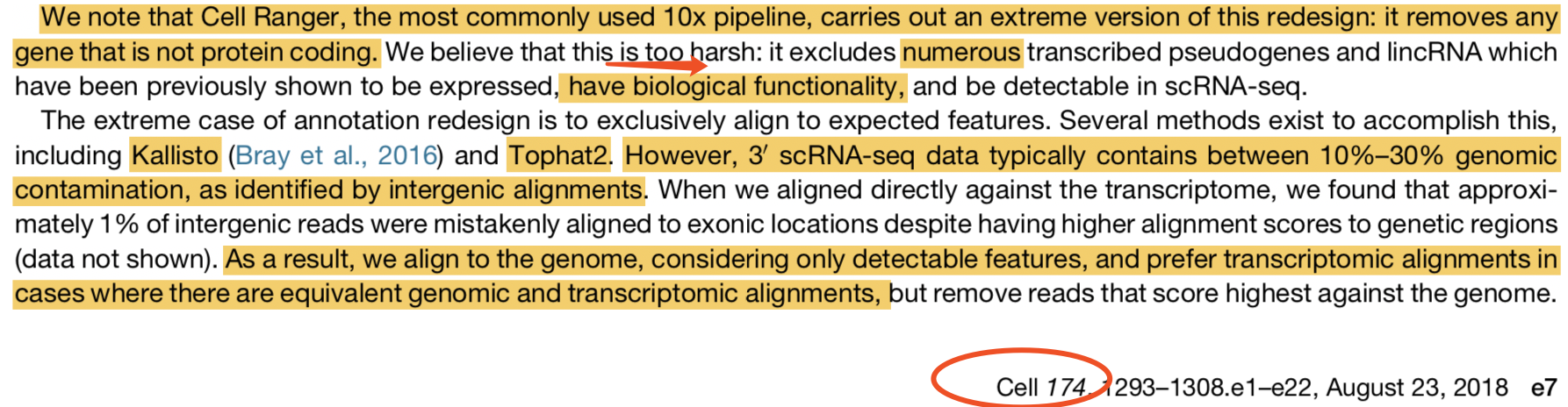
也就是说,作者认为,这个10X仪器的单细胞转录组数据走cellranger流程,其实是有一点问题的。
如果你关心的是走cellranger流程本身,我们在单细胞天地多次分享过流程笔记: - 单细胞实战(一)数据下载
- 单细胞实战(二) cell ranger使用前注意事项
- 单细胞实战(三) Cell Ranger使用初探
- 单细胞实战(四) Cell Ranger流程概览
- 单细胞实战(五) 理解cellranger count的结果
拿到表达矩阵后再走Seurat流程哦,两个流程基本上解决了10X仪器的单细胞转录组数据分析的80%基础内容。作者的解决方案
首先自定义STAR比对软件参数
作者自己使用了STAR比对10X仪器的单细胞转录组数据,其实就是走cellranger流程,因为cellranger里面封装的就是STAR比对软件,但是作者参数选择是:Alignment parameters used are as follows: —outFilterType BySJout, —outFilterMultimapNmax 100, —limitOutSJcollapsed 2000000 —alignSJDBoverhangMin 8, —outFilterMismatchNoverLmax 0.04, – alignIntronMin 20, —alignIntronMax 1000000, —readFilesIn fastqrecords, —outSAMprimaryFlag AllBestScore, —outSAMtype BAM Unsorted
也就是说,作者允许每个测序的reads可以有多达20个位置的比对情况输出,而且没有成功比对的reads也会输出一条记录。然后修复那些多比对情况的reads
既然作者允许每个测序的reads可以有多达20个位置的比对情况输出,那么就得给这些测序的reads一个归属。因为10X仪器的单细胞转录组数据本来就只有3端,而3端的序列本来就是比参考基因组其它区域更加同源。作者考虑了Kallisto (Bray et al., 2016) and EM approaches, such as RSEM (Li and Dewey, 2011), 两个方法,但是后面自己开发的方法有点复杂:

如果你也有兴趣,可以细细品读。我们的介绍就到此为止啦。