前面我们在生信技能树系统性介绍了大量RNA-seq相关背景知识,以及表达矩阵分析的一般流程
- RNA-seq这十年(3万字长文综述)
- RNA-seq的counts值,RPM, RPKM, FPKM, TPM 的异同
- 表达矩阵的归一化和标准化,去除极端值,异常值
- 箱线图的生物学含义
- 转录组表达矩阵为什么需要主成分分析以及怎么做
- limma/voom,edgeR,DESeq2分析注意事项,差异分析表达矩阵与分组信息
其中差异分析我们使用了limma/voom,edgeR,DESeq2这3个流程,很多朋友比较感兴趣到底应该是选择哪一个,而且它们的区别是?
具体的统计学原理我们推荐大家看:
这里我们直接看效果,正好最近重新复习TCGAbiolinks看到了这个图。
学徒作业,完成两个火山图,一个logFC的散点图,一个UpSet图
步骤分解:
- 从UCSC的XENA数据库里面下载TCGA-BRCA 的counts值矩阵
- 从UCSC的XENA数据库里面下载TCGA-BRCA 的亚型信息
- 使用 limma or edgeR 对下载的counts值矩阵根据亚型信息进行差异分析
- 差异分析结果做火山图
- 差异分析结果做logFC的散点图
- 差异分析结果做UpSet图
DEA analyses of TCGA-BRCA data comparing luminal subtypes with normal samples. A-B) Volcano plots are shown where only those genes with logFC higher than 6 or lower than-6 are labelled and only the significant up-or down-regulated genes are shown as dots.
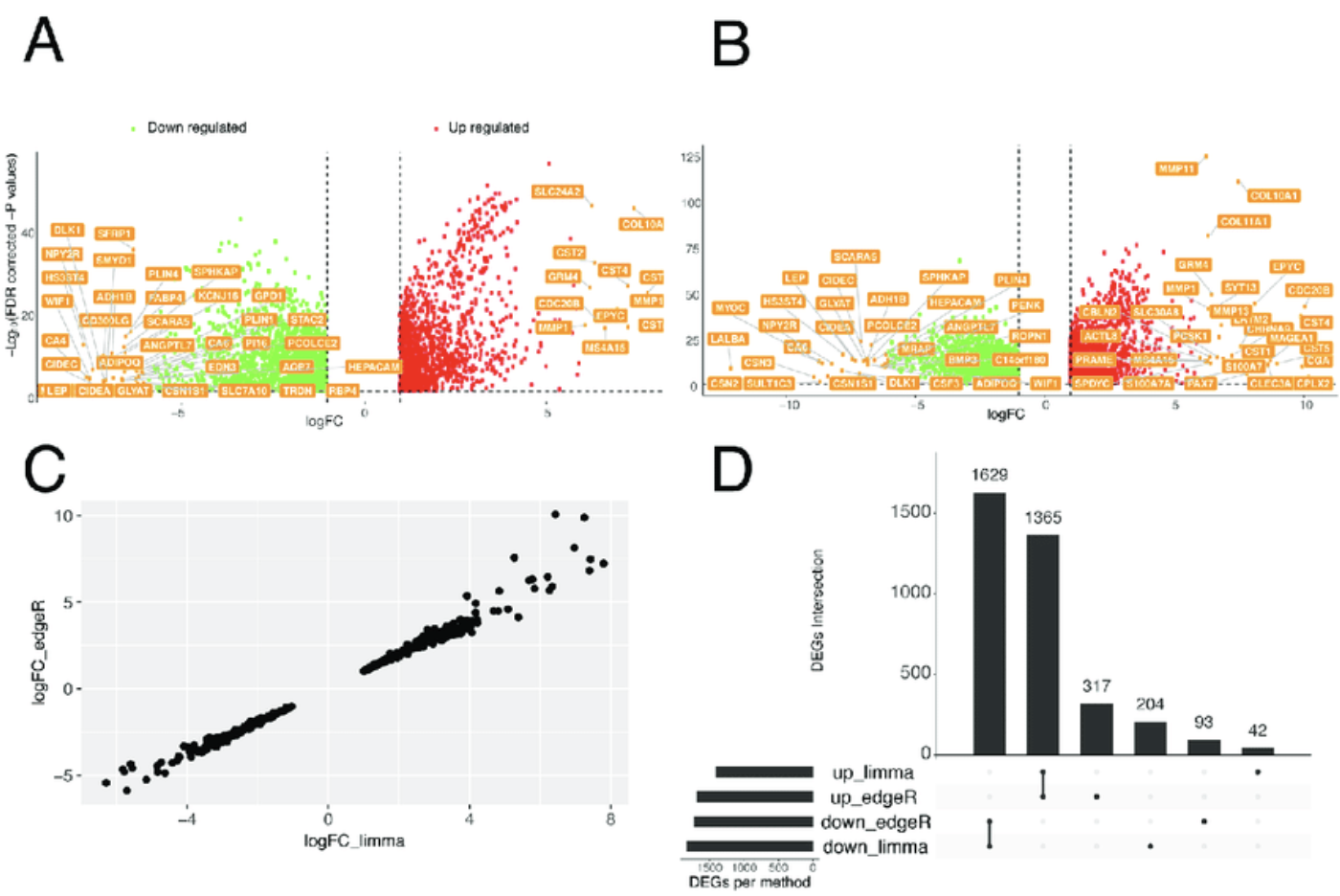
We carried out DEA using the limma (A) or edgeR pipelines (B) of TCGAbiolinks. C) The correlation plot between the logFC estimated by the two pipelines for the top 500 DE genes is shown. The genes discussed in the main text are highlighted in bold. D) The intersect between all the DE genes estimated by the two pipelines is shown using UpSet. https://doi.org/10.1371/journal.pcbi.1006701.g003
TCGAbiolinks值得大家仔细学习
现在的CNS文章,或多或少使用一些TCGA教程,你把这个R包学习10遍,写成200篇自己的笔记,未来一年的学习计划。
这个包涵盖了TCGA的方方面面:https://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html
| HTML | R Script | 1. Introduction |
|---|---|---|
| HTML | R Script | 10. TCGAbiolinks_Extension |
| HTML | R Script | 2. Searching GDC database |
| HTML | R Script | 3. Downloading and preparing files for analysis |
| HTML | R Script | 4. Clinical data |
| HTML | R Script | 5. Mutation data |
| HTML | R Script | 6. Compilation of TCGA molecular subtypes |
| HTML | R Script | 7. Analyzing and visualizing TCGA data |
| HTML | R Script | 8. Case Studies |
| HTML | R Script | 9. Graphical User Interface (GUI) |