是我太年轻
学员群有咨询 Agilent-038314 CBC Homo sapiens lncRNA + mRNA microarray V2.0 这个表达量芯片的数据处理问题,当然了,主要是芯片的探针ID对应基因名字的问题。 链接是;https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL18109
因为大家还是初学者,所以我就想着先打击一下,说这样的芯片比较难,肯定是很少有人挖掘它,因为它仅仅是提供了探针的碱基序列,有一个费时费力的流程去拿到该芯片的注释信息:
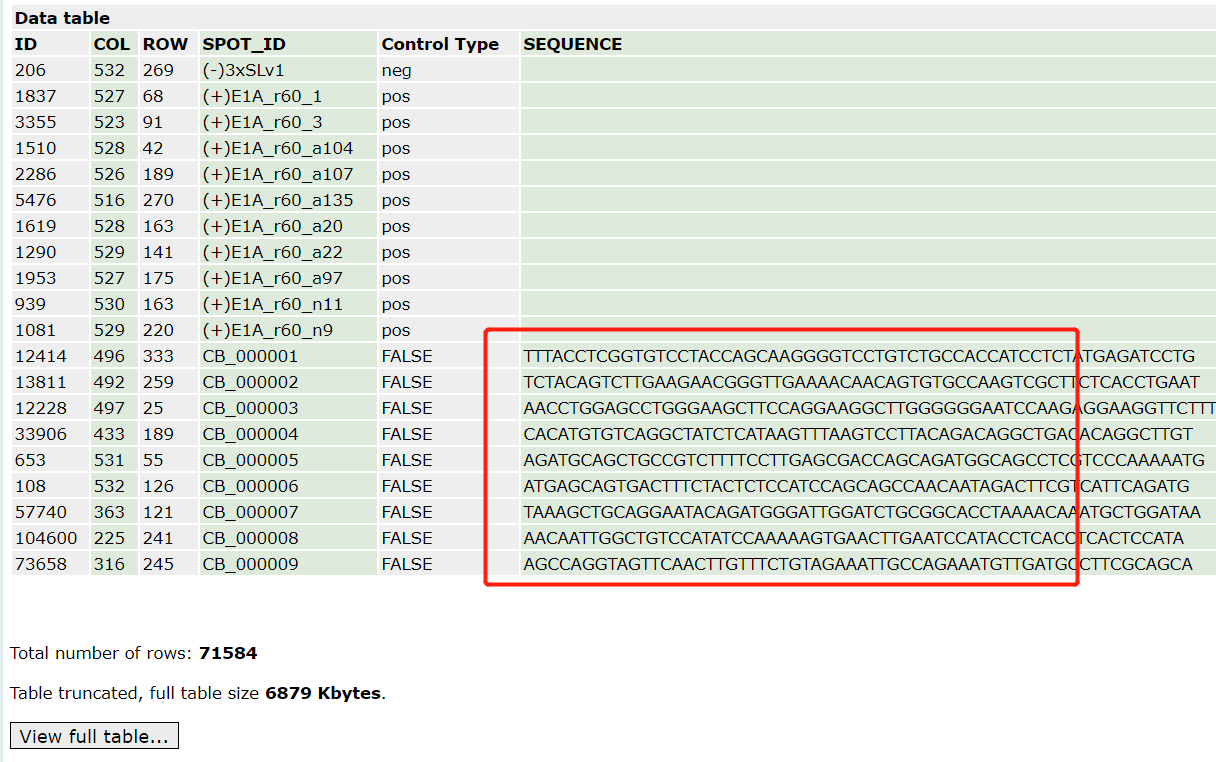
比如我们可以看到《LncRNA and mRNA integration network reconstruction reveals novel key regulators in esophageal squamous-cell carcinoma》 这个2019的文献,链接是: https://doi.org/10.1016/j.ygeno.2018.01.003
就对这个芯片做了非常复杂的处理:

Microarray data contained 71,584 probes. After applying the criteria for re-annotation, 39,068 of the probes were retained, among which 20,323 were corresponding to mRNAs and 18,745 to lncRNA.
最后这些探针还需要去冗余,得到:These probes were mapped to 25,018 unique genes, including 13,490 protein coding genes (PCGs) and 11,528 lncRNAs.
而且这些数据,文章都整理好了,都在附件:
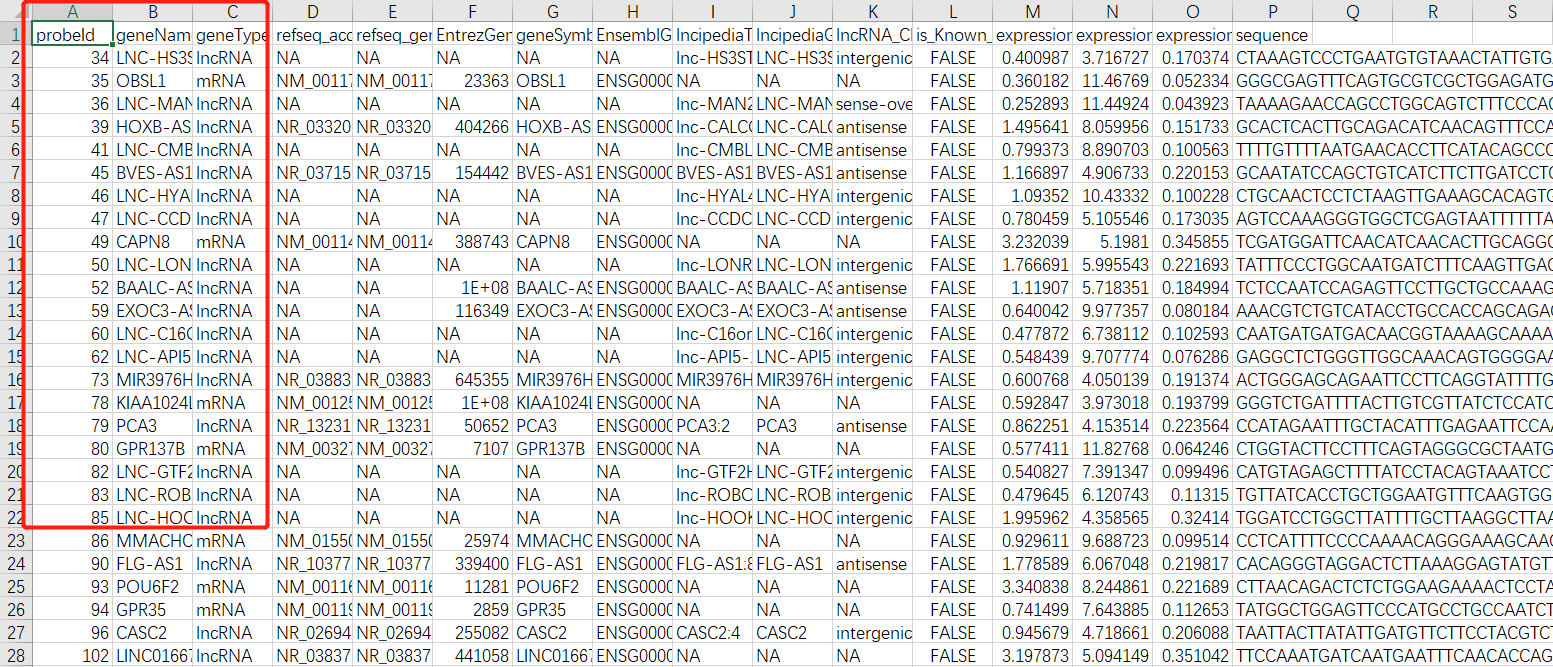
现在你还在发愁,这样的芯片,如何做ID转化吗?
让我吃惊的是
出于职业习惯,我去看了看这个数据集页面: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53625
发现它居然链接到了4个文献:
- Li J, Chen Z, Tian L, Zhou C et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut 2014 Nov;63(11):1700-10. PMID: 24522499
- Li W, Zhang L, Guo B, Deng J et al. Exosomal FMR1-AS1 facilitates maintaining cancer stem-like cell dynamic equilibrium via TLR7/NFκB/c-Myc signaling in female esophageal carcinoma. Mol Cancer 2019 Feb 8;18(1):22. PMID: 30736860
- Li Y, Lu Z, Che Y, Wang J et al. Immune signature profiling identified predictive and prognostic factors for esophageal squamous cell carcinoma. Oncoimmunology 2017;6(11):e1356147. PMID: 29147607
- Shi X, Chen Z, Hu X, Luo M et al. AJUBA promotes the migration and invasion of esophageal squamous cell carcinoma cells through upregulation of MMP10 and MMP13 expression. Oncotarget 2016 Jun 14;7(24):36407-36418. PMID: 27172796
- Liu J, Wang Y, Chu Y, Xu R et al. Identification of a TLR-Induced Four-lncRNA Signature as a Novel Prognostic Biomarker in Esophageal Carcinoma. Front Cell Dev Biol 2020;8:649. PMID: 32850794
我又好奇去谷歌搜了一下这个数据集:
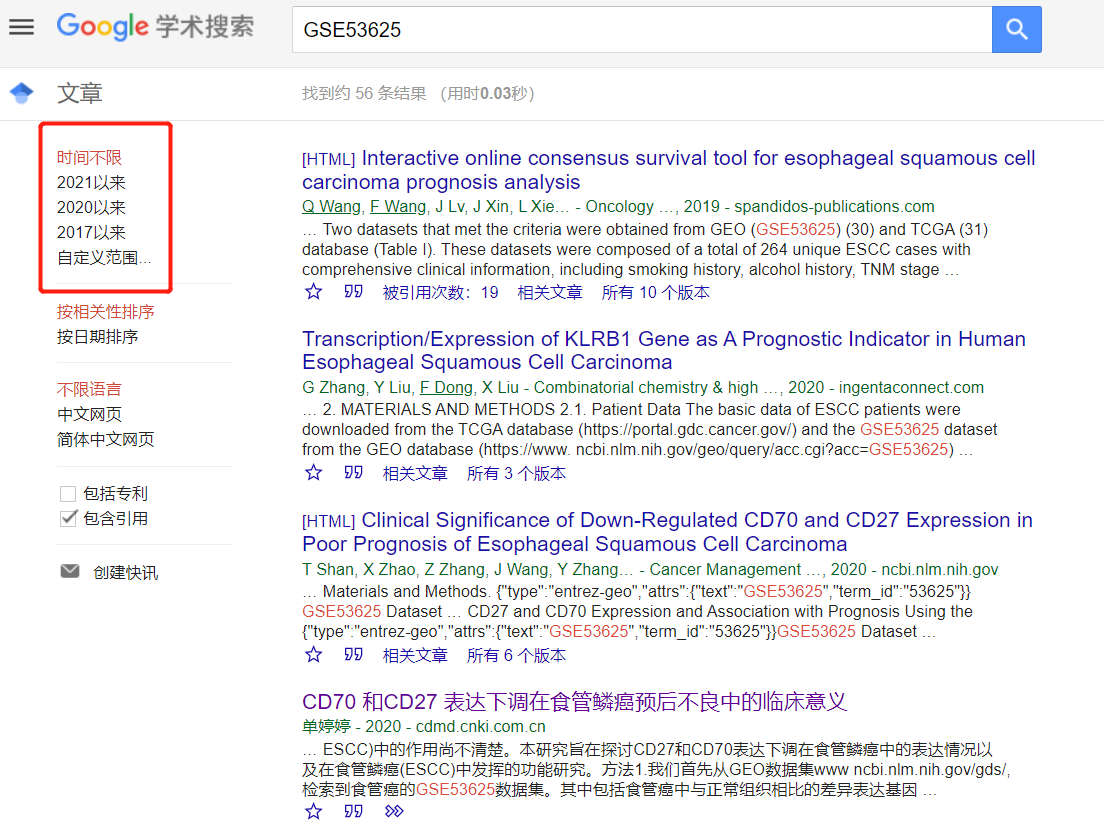
粗略看了看,起码几十篇数据挖掘文献, 就针对这一个数据集,变着花样各种挖,活脱脱的一个数据挖掘发展史!
常规的差异分析呢,基本上看我六年前的表达芯片的公共数据库挖掘系列推文即可; - 解读GEO数据存放规律及下载,一文就够
- 解读SRA数据库规律一文就够
- 从GEO数据库下载得到表达矩阵 一文就够
- GSEA分析一文就够(单机版+R语言版)
- 根据分组信息做差异分析- 这个一文不够的
- 差异分析得到的结果注释一文就够
文末友情推荐
- 学徒培养2021名额开放申请
- 老板,请为我配备一个懂生信的师兄
- 你以为GEO只是挖挖就完了吗