最近刷到了2023发表在NC杂志的男性乳腺癌患者的单细胞转录组图谱文章,标题是:《Single-cell transcriptome analysis indicates fatty acid metabolism-mediated metastasis and immunosuppression in male breast cancer》
其中附件有一张图是男性和女性的乳腺癌患者肿瘤细胞表达量差异基因的代谢通路打分后的差异热图,如下所示:
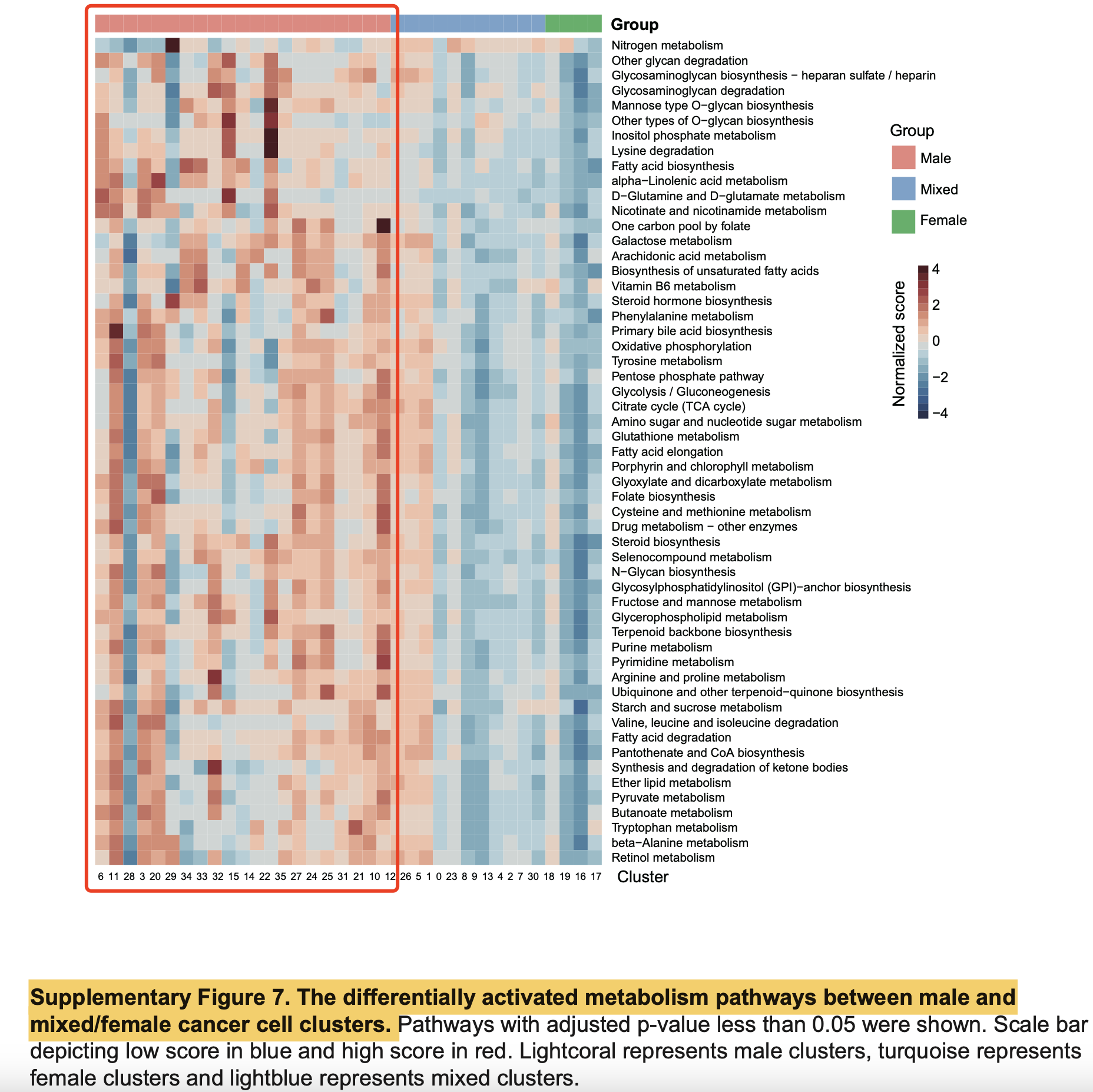
可以看到有50多个代谢通路都是在男性乳腺癌患者里面的癌细胞是显著的激活相当于女性乳腺癌患者来说,略微有那么一点点不可思议了哦。
我们来一个学徒作业吧,大家可以针对常见的性别不显著的癌症去搜索单细胞转录组数据集,里面肯定是有一些病人性别信息的,就可以做类似的分析看看。
首先获取在kegg数据库的全部的代谢通路
library(KEGGREST)
org <- keggList('organism')
head(org)
org[str_detect(org[,3],"human"),]
hsa_path <- keggLink("pathway","hsa")
length(hsa_path)
length(unique(names(hsa_path)))
length(unique(hsa_path))
# 2024-05-30 19:17:33
# kegg数据库目前记录了 8779 个基因
# kegg数据库目前记录了 359 个通路
# 它们之间的组合是 36981
unique(hsa_path)
# 其中 hsa00 开头的就是代谢相关
index <- grepl('hsa00',unique(hsa_path))
meta= unique(hsa_path)[index]
meta
hsa_info <- lapply(meta, keggGet)
# 获取代谢通路名字
nm=unlist(lapply( hsa_info , function(x) x[[1]]$NAME))
nm
可以看到是如下所示的 84个通路 :
[1] "Glycolysis / Gluconeogenesis"
[2] "Citrate cycle (TCA cycle)"
[3] "Pentose phosphate pathway"
[4] "Pentose and glucuronate interconversions"
[5] "Fructose and mannose metabolism"
[6] "Galactose metabolism"
[7] "Ascorbate and aldarate metabolism"
[8] "Fatty acid biosynthesis"
[9] "Fatty acid elongation"
[10] "Fatty acid degradation"
[11] "Steroid biosynthesis"
[12] "Primary bile acid biosynthesis"
[13] "Ubiquinone and other terpenoid-quinone biosynthesis"
[14] "Steroid hormone biosynthesis"
[15] "Oxidative phosphorylation"
[16] "Arginine biosynthesis"
[17] "Purine metabolism"
[18] "Caffeine metabolism"
[19] "Pyrimidine metabolism"
[20] "Alanine, aspartate and glutamate metabolism"
[21] "Glycine, serine and threonine metabolism"
[22] "Cysteine and methionine metabolism"
[23] "Valine, leucine and isoleucine degradation"
[24] "Valine, leucine and isoleucine biosynthesis"
[25] "Lysine degradation"
[26] "Arginine and proline metabolism"
[27] "Histidine metabolism"
[28] "Tyrosine metabolism"
[29] "Phenylalanine metabolism"
[30] "Tryptophan metabolism"
[31] "Phenylalanine, tyrosine and tryptophan biosynthesis"
[32] "beta-Alanine metabolism"
[33] "Taurine and hypotaurine metabolism"
[34] "Phosphonate and phosphinate metabolism"
[35] "Selenocompound metabolism"
[36] "D-Amino acid metabolism"
[37] "Glutathione metabolism"
[38] "Starch and sucrose metabolism"
[39] "N-Glycan biosynthesis"
[40] "Other glycan degradation"
[41] "Mucin type O-glycan biosynthesis"
[42] "Various types of N-glycan biosynthesis"
[43] "Other types of O-glycan biosynthesis"
[44] "Mannose type O-glycan biosynthesis"
[45] "Amino sugar and nucleotide sugar metabolism"
[46] "Neomycin, kanamycin and gentamicin biosynthesis"
[47] "Glycosaminoglycan degradation"
[48] "Glycosaminoglycan biosynthesis - chondroitin sulfate / dermatan sulfate"
[49] "Glycosaminoglycan biosynthesis - keratan sulfate"
[50] "Glycosaminoglycan biosynthesis - heparan sulfate / heparin"
[51] "Glycerolipid metabolism"
[52] "Inositol phosphate metabolism"
[53] "Glycosylphosphatidylinositol (GPI)-anchor biosynthesis"
[54] "Glycerophospholipid metabolism"
[55] "Ether lipid metabolism"
[56] "Arachidonic acid metabolism"
[57] "Linoleic acid metabolism"
[58] "alpha-Linolenic acid metabolism"
[59] "Sphingolipid metabolism"
[60] "Glycosphingolipid biosynthesis - lacto and neolacto series"
[61] "Glycosphingolipid biosynthesis - globo and isoglobo series"
[62] "Glycosphingolipid biosynthesis - ganglio series"
[63] "Pyruvate metabolism"
[64] "Glyoxylate and dicarboxylate metabolism"
[65] "Propanoate metabolism"
[66] "Butanoate metabolism"
[67] "One carbon pool by folate"
[68] "Thiamine metabolism"
[69] "Riboflavin metabolism"
[70] "Vitamin B6 metabolism"
[71] "Nicotinate and nicotinamide metabolism"
[72] "Pantothenate and CoA biosynthesis"
[73] "Biotin metabolism"
[74] "Lipoic acid metabolism"
[75] "Folate biosynthesis"
[76] "Retinol metabolism"
[77] "Porphyrin metabolism"
[78] "Terpenoid backbone biosynthesis"
[79] "Nitrogen metabolism"
[80] "Sulfur metabolism"
[81] "Aminoacyl-tRNA biosynthesis"
[82] "Metabolism of xenobiotics by cytochrome P450"
[83] "Drug metabolism - cytochrome P450"
[84] "Drug metabolism - other enzymes"
其实这些代谢通路还是可以分类的,在 https://www.genome.jp/kegg-bin/show_organism?menu_type=pathway_maps&org=hsa 可以看到,有几个代谢通路居然是例外,并不是00开头,都属于Global and overview maps,如下所示:
Global and overview maps
01100 Metabolic pathways
01200 Carbon metabolism
01210 2-Oxocarboxylic acid metabolism
01212 Fatty acid metabolism
01230 Biosynthesis of amino acids
01232 Nucleotide metabolism
01250 Biosynthesis of nucleotide sugars
01240 Biosynthesis of cofactors
比如我们使用y叔的包试试看:
library(clusterProfiler)
library(ggplot2)
data(geneList, package='DOSE')
de <- names(geneList)[1:1000]
yy <- enrichKEGG(de, pvalueCutoff=1,
qvalueCutoff = 1)
head(yy@result[,1:3])
dim(yy@result)
tmp=yy@result
colnames(tmp)
sort(table(tmp$subcategory[tmp$category=='Metabolism']))
可以看到有73个代谢通路,如下所示的分类 :
> as.data.frame(sort(table(tmp$subcategory[tmp$category=='Metabolism'])))
Var1 Freq
1 Metabolism of terpenoids and polyketides 1
2 Nucleotide metabolism 2
3 Energy metabolism 3
4 Metabolism of other amino acids 3
5 Xenobiotics biodegradation and metabolism 3
6 Global and overview maps 6
7 Metabolism of cofactors and vitamins 8
8 Amino acid metabolism 9
9 Glycan biosynthesis and metabolism 10
10 Carbohydrate metabolism 12
11 Lipid metabolism 13
另外就是 org.Hs.eg.db 里面也存储了kegg数据库信息,但是非常老旧了,可以看到:
library(org.Hs.eg.db)
ids=toTable(org.Hs.egPATH)
head(ids)
length(unique(ids$path_id))