安排学徒复现一个新鲜出炉的阿兹海默症的单细胞文章:《Characterisation of premature cell senescence in Alzheimer’s disease using single nuclear transcriptomics》:
- Acta Neuropathologica (2024)
- https://doi.org/10.1007/s00401-024-02727-9
对应的数据集是:GSE264648,大家很容易读取作者提供的表达量矩阵然后进行降维聚类分群。值得注意的是作者重点看了看 the ‘canonical senescence pathway (CSP)’, the ‘senescence initiating pathway (SIP)’ 这两个基因列表在单细胞亚群的差异情况,如下所示:
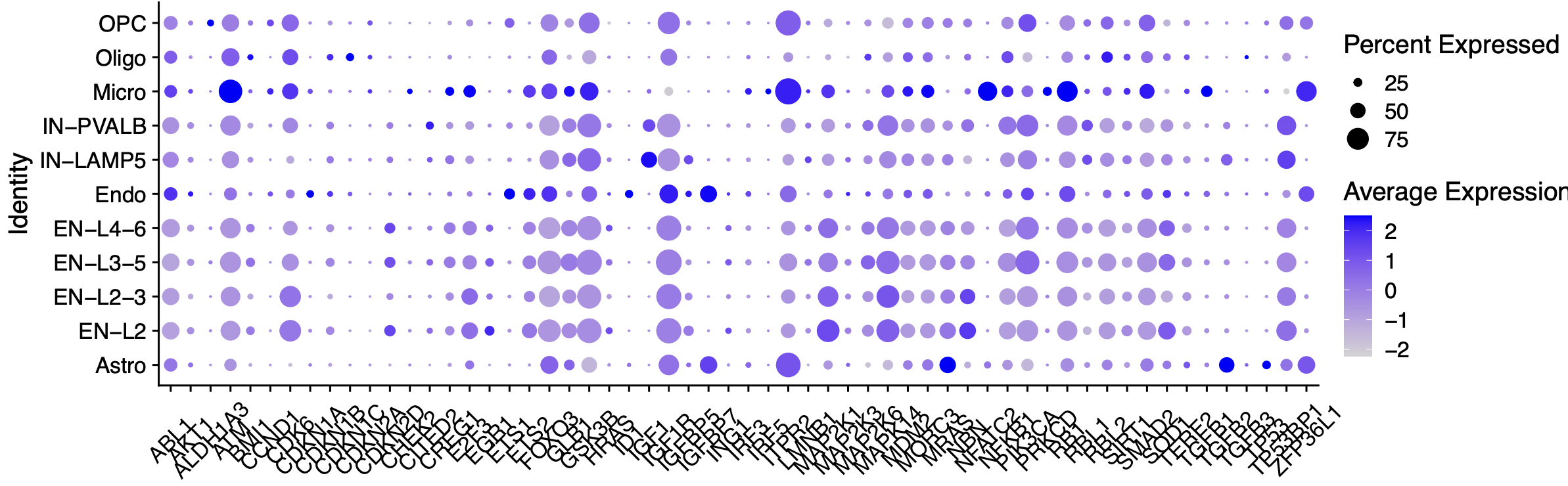
左边的气泡图很容易理解,也很容易复现,在前面的降维聚类分群结果的基础上面进行:
# 创建基因向量
gene_vector <- c("ABL1", "AKT1", "ALDH1A3", "ATM", "BMI1", "CCND1", "CDK6",
"CDKN1A", "CDKN1B", "CDKN1C", "CDKN2A", "CDKN2D", "CHEK2", "CITED2",
"CREG1", "E2F3", "EGR1", "ETS1", "ETS2", "FOXO3", "GLB1", "GSK3B",
"HRAS", "ID1", "IGF1", "IGF1R", "IGFBP5", "IGFBP7", "ING1", "IRF3",
"IRF5", "ITPR2", "LMNB1", "MAP2K1", "MAP2K3", "MAP2K6", "MAPK14",
"MDM2", "MORC3", "MRAS", "NBN", "NFATC2", "NFKB1", "PIK3CA", "PRKCD",
"RB1", "RBL1", "RBL2", "SIRT1", "SMAD2", "SOD1", "TERF2", "TGFB1",
"TGFB2", "TGFB3", "TP53", "TP53BP1", "ZFP36L1")
# 打印基因向量
print(gene_vector)
p2 = DotPlot( sce.all.int, features = gene_vector,
group.by = 'celltype') +
theme(axis.text.x = element_text(angle = 45, vjust = 0.5, hjust=0.5))
p2
ggsave('DotPlot-for-brain-senescence.pdf',width=12)
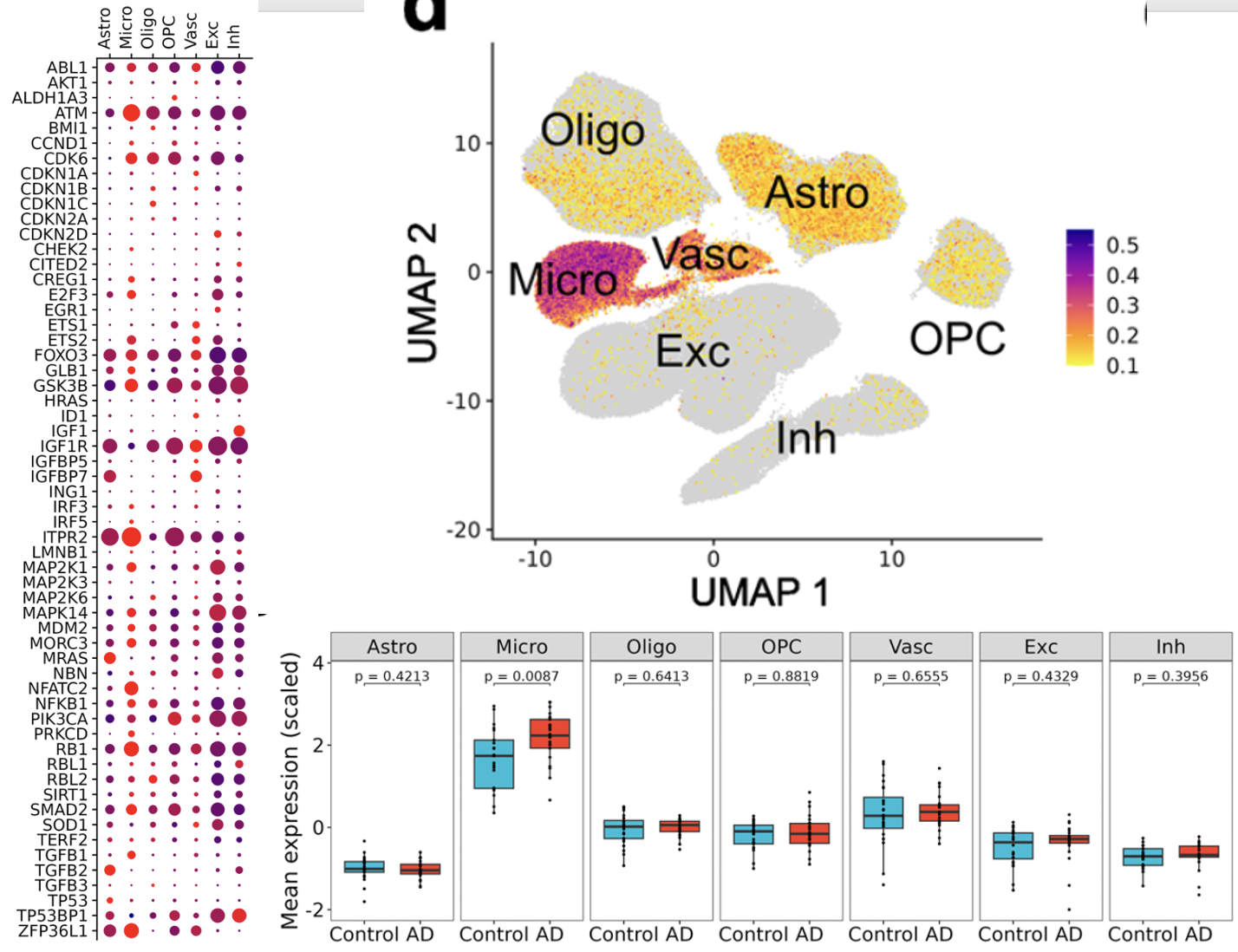
就可以拿到了几乎是一模一样的图,而且是肉眼都可以看到在小胶质细胞和内皮细胞是有一些高表达量的趋势 :
但是这些基因如何汇总成为一个一个Normalised aggregated expression 后,然后projected on UMAP上面呢,文章写的是
Gene set or module feature plots were generated by first calculating aggregated gene set scores using AddModuleScore() function from Seurat and then plotted using FeaturePlot scCustom() from scCustomize
我这里测试了UCell的打分, 因为它计算会比较快 :
# remotes::install_github("carmonalab/UCell")
library(UCell)
scRNA=sce.all.int
gene_vector=list(gene_vector=gene_vector)
gene_vector
names(gene_vector)
sc_dataset <- AddModuleScore_UCell(scRNA,
features = gene_vector)
signature.names <- paste0(names(gene_vector), "_UCell")
options(repr.plot.width=6, repr.plot.height=4)
colnames(sc_dataset@meta.data)
VlnPlot(sc_dataset, features = signature.names,
group.by = "celltype",pt.size = 0 ) + NoLegend()
ggsave('VlnPlot-senescence.pdf',width = 7)
FeaturePlot(sc_dataset,'gene_vector_UCell')
ggsave('FeaturePlot-senescence.pdf',width = 5)
table(sc_dataset$batch)
sc_dataset$diagnosis= ifelse(grepl('AD',sc_dataset$batch),'AD','control')
VlnPlot(sc_dataset, features = signature.names,
split.by = 'diagnosis',
group.by = "celltype",pt.size = 0 ) + NoLegend()
ggsave('VlnPlot-senescence-diagnosis.pdf',width = 7)
FeaturePlot(sc_dataset,
split.by = 'diagnosis',
'gene_vector_UCell')
ggsave('FeaturePlot-senescence-diagnosis.pdf',width = 8)
确实是很明显的在小胶质细胞和内皮细胞这个打分高很多,但是这个打分在阿兹海默症和正常组是否有差异就值得商榷了。因为文章里面把打分居然还进一步进行了scale后展示:
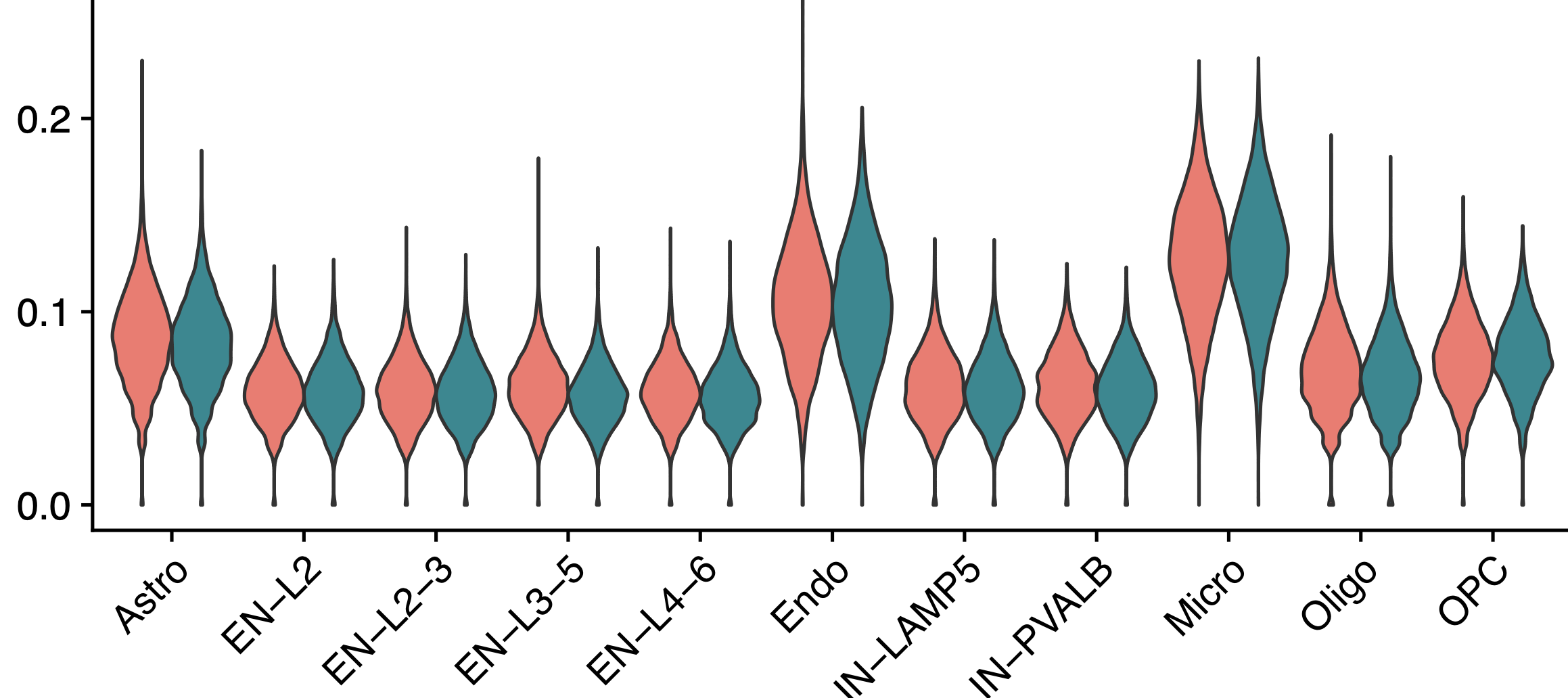
学徒作业
大家试试看这个编号为GSE264648的数据集的降维聚类分群,然后直接使用作者自己的单细胞亚群命名结果,去可视化所谓的the ‘canonical senescence pathway (CSP)’, the ‘senescence initiating pathway (SIP)’ 这两个基因列表的打分情况。
首先看看是不是在小胶质细胞有特异性,另外看看是不是小胶质的这个打分在阿兹海默症和正常组是否有差异!