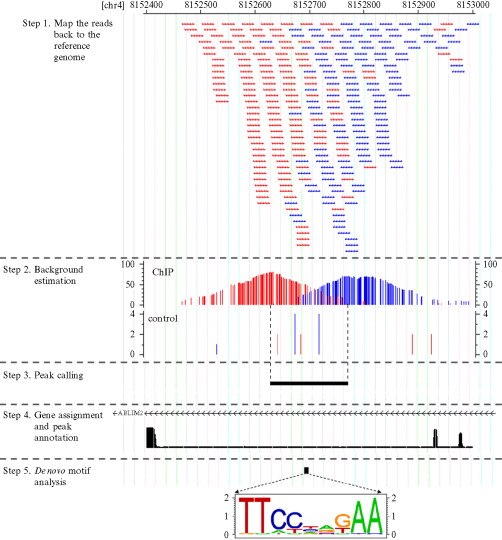
我只能说,CHIP-seq的确是非常完善的NGS流程了,各种资料层出不穷,大家首先可以看下面几个完整流程的PPT来对CHIP-seq流程有个大致的印象,我对前面提到的文献数据处理的几个要点,就跟下面这个图片类似:
QuEST is a statistical software for analysis of ChIP-Seq data with data and analysis results visualization through UCSC Genome Browser. http://www-hsc.usc.edu/~valouev/QuEST/QuEST.html
peak calling 阈值的选择: http://www.nature.com/nprot/journal/v7/n1/fig_tab/nprot.2011.420_F2.html
MeDIP-seq and histone modification ChIP-seq analysis http://crazyhottommy.blogspot.com/2014/01/medip-seq-and-histone-modification-chip.html
2011-review-CHIP-seq-high-quaility-data: http://www.nature.com/ni/journal/v12/n10/full/ni.2117.html?message-global=remove
不同处理条件的CHIP-seq的差异peaks分析: http://www.slideshare.net/thefacultyl/diffreps-automated-chipseq-differential-analysis-package
一个实际的CHIP-seq数据分析例子: http://www.biologie.ens.fr/~mthomas/other/chip-seq-training/
然后下面的各种资料,是针对CHIP-seq流程的各个环境的,还有一些是针对于表观遗传学知识的
## pipeline : https://github.com/shenlab-sinai/chip-seq_preprocess
## https://sites.google.com/site/anshul...e/projects/idr ## samtools view -b -F 1548 -q 30 chipSampleRep1.bam
## Hands-on introduction to ChIP-seq analysis - VIB Training http://www.biologie.ens.fr/~mthomas/other/chip-seq-training/
## video(A Step-by-Step Guide to ChIP-Seq Data Analysis Webinar) : http://www.abcam.com/webinars/a-step-by-step-guide-to-chip-seq-data-analysis-webinar
## Using ChIP-Seq to identify and/or quantify bound regions (peaks) http://barcwiki.wi.mit.edu/wiki/SOPs/chip_seq_peaks
## 公开课: https://www.coursera.org/learn/galaxy-project/lecture/FUzcg/chip-sequence-analysis-with-macs
## 日语教程:http://genomejack.net/download/GenomeJackBrowserAppendix/browser_appendix_j/tutorials/chipSeq.html
## 台湾教程:http://lsl.sinica.edu.tw/Services/Class/files/20151118475_2.pdf 徐唯哲 Paul Wei-Che HSU
中央研究院 分子生物研究所
研究助技師
## peak finder软件大全: http://wodaklab.org/nextgen/data/peakfinders.html
## paper: Large-Scale Quality Analysis of Published ChIP-seq Data http://www.g3journal.org/content/4/2/209.full
## paper: Chip-seq data analysis: from quality check to motif discovery and more http://ccg.vital-it.ch/var/sib_april15/cases/landt12/strand_correlation.html
## Workshop hands on session(RNA-Seq / ChIP-Seq ) : https://hpc.oit.uci.edu/biolinux/handson.docx
## paper supplement : http://genome.cshlp.org/content/suppl/2015/10/02/gr.192005.115.DC1/Supplemental_Information.docx
http://www.ncbi.nlm.nih.gov/pubmed/22130887 "Analyzing ChIP-seq data: preprocessing, normalization, differential identification, and binding pattern characterization."
http://www.ncbi.nlm.nih.gov/pubmed/22499706 "Normalization, bias correction, and peak calling for ChIP-seq." (stat heavy)
http://www.ncbi.nlm.nih.gov/pubmed/24244136 "Practical guidelines for the comprehensive analysis of ChIP-seq data."
http://www.ncbi.nlm.nih.gov/pubmed/25223782 "Identifying and mitigating bias in next-generation sequencing methods for chromatin biology."
A quick search also turned up this recent paper (which I haven't read) that might be of interest to you
http://www.ncbi.nlm.nih.gov/pubmed/24598259 "Impact of sequencing depth in ChIP-seq experiments."
## figures: https://github.com/shenlab-sinai/ngsplot
还有两个web-tools也是可视化
bioconductor系列工具和教程 :
http://faculty.ucr.edu/~tgirke/HTML_Presentations/Manuals/Workshop_Dec_6_10_2012/Rchipseq/Rchipseq.pdf