主要是做测序
To decipher the molecular pathogenesis of paediatric GBM, we undertook a comprehensive mutation analysis in protein-coding genes by performing whole-exome sequencing (WES) on 48 well-characterized paediatric GBMs, including 6 patients for whom we had matched non-tumour (germline) DNA.
只有6个患者有NT配对样本,用来找somatic的mutation,结果发现其中4个患者就有H3F3A的突变,但是H3F3A本身是非常保守的,所以这个现象值得研究。
to our knowledge no human disorders have specifically been associated with mutations in histones, including H3.3
所以才扩大了WES测序样本数量。
(数据在 https://www.ebi.ac.uk/ega/studies/EGAS00001000226 )
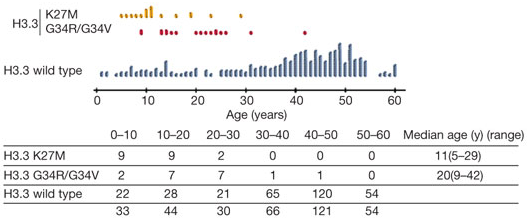
结果:A total of 15 samples had heterozygous H3.3 mutations (9 K27M, 5 G34R, 1 G34V) and 14 samples had a mutation in ATRX
但是早先的TCGA计划中的GBM只测序了600个基因,不包括我们感兴趣的基因,而且公共数据里面的22个成人GBM里面 没有发现H3F3A相关突变。
所以把自己医院的所有样本(784 gliomas)都测了个遍,总结如下:
因为H3F3A突变了,同时需要探究
ATRX, which encodes a member of a transcription/chromatin remodelling complex required for the incorporation of H3.3 at pericentric heterochromatin and at telomeres, as well as at several transcription factor binding sites
DAXX (because the gene product heterodimerizes with ATRX and participates in H3.3 recruitment to DNA
同时也探究了27个样本的mRNA表达水平
showed a clear mutation-specific expression pattern when comparing both between K27 and G34 mutants and with H3.3 wild-type GBMs, including DLX2, SFRP2, FZD7 and MYT1
数据在; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE34824
RNA from 27 primary tumor samples was subject to QC by Agilent BioAnalyser analysis and then hybridised to an Affymetrix U133 Plus2 gene expression array
同时也探究了拷贝数:
DNA from 31 of the 48 paediatric GBM tumours analysed by whole-exome sequencing was hybridized to Illumina Human Omni 2.5M Single Nucleotide Polymorphism (SNP) arrays
但是这个数据无法找到。