每次开展R语言线下学习班,都需要重新发几次:Windows电脑使用Rstudio会有多少错误呢 ,虽然大部分同学都是可以根据我们的教程顺利解决问题,但是不幸的人各有各的不幸。一般来说就是Windows电脑的中文用户名需要修改电脑系统的环境变量,R包下载等等。
但是今天有一个学员起初是下载R包无法联网,所以失败,根据我们的经验当然是options(download.file.method = ‘libcurl’)就轻轻松松解决啦,不过这次居然是仅仅是解决了R自带R包下载问题,使用BiocManager仍然是无法安装R包,如下所示:
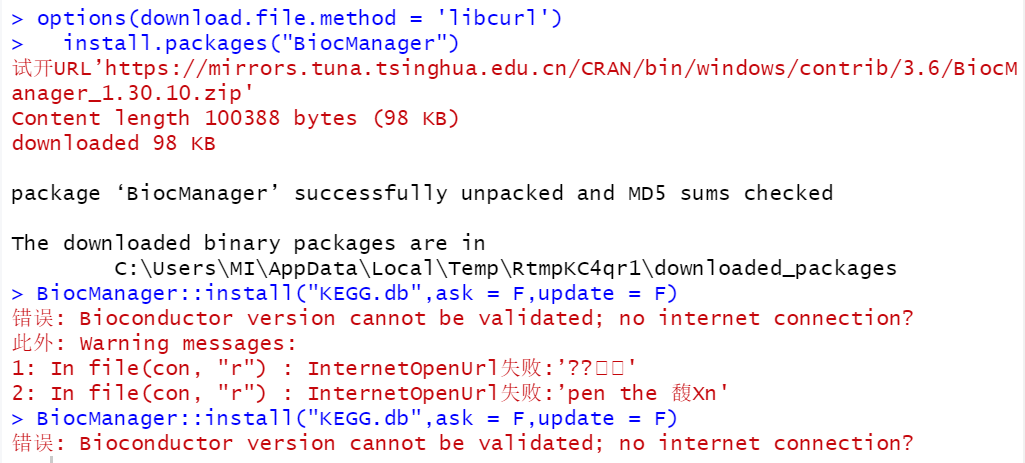
当然就得根据关键词去搜索啦!
> BiocManager::install("KEGG.db",ask = F,update = F)
错误: Bioconductor version cannot be validated; no internet connection?
此外: Warning messages:
第一次搜索以为是http和https的问题
参考:https://stackoverflow.com/questions/33355444/r-when-trying-to-install-package-internetopenurl-failed
大概意思是让我修改镜像把所有的https都替换为http,步骤如下:
- using regular http mirrors instead of https
- update your CA certificate bundle to allow proper certificate validation
- setting the default download method to “libcurl” and see if that helps: options(download.file.method=”libcurl”)
但是尝试了,失败!第二次尝试以为是BiocManager的config文件
所以根据搜索结果使用:
config <- readLines("https://bioconductor.org/config.yaml")这个时候的报错让我恍然大悟

其实真正的问题还是在联网上面!也就是说options(download.file.method = ‘libcurl’)并没有完全解决问题。配置R联网环境
继续搜索R联网环境,这个时候发现了url.method这个配置的解决方案;
options(download.file.method = 'libcurl') options(url.method='libcurl')果然,现在在Windows电脑里面R语言的安装R包和下载文件就OK啦。有趣的是,你每次都需要复制粘贴上面两行代码,或者你把它写入到你的R配置文件哦。
那么这两个参数到底是啥区别呢
首先看 download.file.method ,非常复杂:https://stat.ethz.ch/R-manual/R-devel/library/utils/html/download.file.htmlMethod to be used for downloading files. Current download methods are "internal", "wininet" (Windows only) "libcurl", "wget" and "curl", and there is a value "auto": see ‘Details’ and ‘Note’. The method can also be set through the option "download.file.method": see options().然后看,也很复杂:https://stat.ethz.ch/R-manual/R-devel/library/base/html/options.html·
character string: the default method for url. Normally unset, which is equivalent to "default", which is "internal" except on Windows.接下来就继续安装R包吧
使用管理员打开R哦,然后就
options()$repos options()$BioC_mirror options(BioC_mirror="https://mirrors.ustc.edu.cn/bioc/") options("repos" = c(CRAN="https://mirrors.tuna.tsinghua.edu.cn/CRAN/")) options()$repos options()$BioC_mirror # https://bioconductor.org/packages/release/bioc/html/GEOquery.html if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager") BiocManager::install("KEGG.db",ask = F,update = F) BiocManager::install(c("GSEABase","GSVA","clusterProfiler" ),ask = F,update = F) BiocManager::install(c("GEOquery","limma","impute" ),ask = F,update = F) BiocManager::install(c("org.Hs.eg.db","hgu133plus2.db" ),ask = F,update = F)如果你还报错,就看看周围有没有大神帮你解决吧!