TCGA计划官方文章在:https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/publications
全部的标题的英文很容易提取和整理,如下:
Comprehensive genomic characterization defines human glioblastoma genes and core pathways
Integrated genomic analyses of ovarian carcinoma
Comprehensive molecular characterization of human colon and rectal cancer
Comprehensive molecular portraits of human breast tumours
Comprehensive genomic characterization of squamous cell lung cancers
Integrated genomic characterization of endometrial carcinoma
Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia
Comprehensive molecular characterization of clear cell renal cell carcinoma
The Cancer Genome Atlas Pan-Cancer analysis project
The somatic genomic landscape of glioblastoma
Comprehensive molecular characterization of urothelial bladder carcinoma
Comprehensive molecular profiling of lung adenocarcinoma
Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin
The Somatic Genomic Landscape of Chromophobe Renal Cell Carcinoma
Comprehensive molecular characterization of gastric adenocarcinoma
Integrated genomic characterization of papillary thyroid carcinoma
Comprehensive genomic characterization of head and neck squamous cell carcinomas
Genomic Classification of Cutaneous Melanoma
Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade Gliomas
Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer
The Molecular Taxonomy of Primary Prostate Cancer
Comprehensive Molecular Characterization of Papillary Renal-Cell Carcinoma
Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma
Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas
Integrated genomic characterization of oesophageal carcinoma
Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma
Integrated Molecular Characterization of Uterine Carcinosarcoma
Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles
Integrated genomic and molecular characterization of cervical cancer
Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma
Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma
Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma
Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer
Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas
The Integrated Genomic Landscape of Thymic Epithelial Tumors
Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas
Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines
Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers
Systematic Analysis of Splice-Site-Creating Mutations in Cancer
Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types
The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma
Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context
Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images
Machine Learning Detects Pan-cancer Ras Pathway Activation in The Cancer Genome Atlas
Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas
Driver Fusions and Their Implications in the Development and Treatment of Human Cancers
Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas
Integrated Genomic Analysis of the Ubiquitin Pathway across Cancer Types
SnapShot: TCGA-Analyzed Tumors
The Cancer Genome Atlas: Creating Lasting Value beyond Its Data
Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation
Oncogenic Signaling Pathways in The Cancer Genome Atlas
Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics
Comprehensive Characterization of Cancer Driver Genes and Mutations
An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics
Pathogenic Germline Variants in 10,389 Adult Cancers
A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples
Genomic and Functional Approaches to Understanding Cancer Aneuploidy
A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers
Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas
lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer
The Immune Landscape of Cancer
Integrated Molecular Characterization of Testicular Germ Cell Tumors
Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients
A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily
Integrative Molecular Characterization of Malignant Pleural Mesothelioma
The chromatin accessibility landscape of primary human cancers
Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer
Before and After: Comparison of Legacy and Harmonized TCGA Genomic Data Commons’ Data
Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer
简单的使用bing搜索一下关键词:word clound in r ,就可以找到解决方案,第一个链接就是:http://www.sthda.com/english/wiki/text-mining-and-word-cloud-fundamentals-in-r-5-simple-steps-you-should-know,代码分成5个步骤。
- Step 1: Create a text file
- Step 2 : Install and load the required packages
- Step 3 : Text mining
- Step 4 : Build a term-document matrix
- Step 5 : Generate the Word cloud
一般来说,会R基础的朋友们很容易看懂,如果你还不会R语言,建议看:
把R的知识点路线图搞定,如下:
- 了解常量和变量概念
- 加减乘除等运算(计算器)
- 多种数据类型(数值,字符,逻辑,因子)
- 多种数据结构(向量,矩阵,数组,数据框,列表)
- 文件读取和写出
- 简单统计可视化
- 无限量函数学习
核心代码就是wordcloud函数,但是这个wordcloud函数要求的输入数据就需要认真做出来。
# 安装R包相信无需再强调了
library("tm")
library("SnowballC")
library("wordcloud")
library("RColorBrewer")
# 这里我们直接读取自己电脑剪切的数据即可
# 运行下面这句代码的同时,需要保证你已经复制了前面我们整理好的文章标题哦!
text=readLines(pipe("pbpaste"))
# 好像这里Mac系统跟Windows系统稍微不一样,大家需要自行把握
# Load the data as a corpus
docs <- Corpus(VectorSource(text))
toSpace <- content_transformer(function (x , pattern ) gsub(pattern, " ", x))
docs <- tm_map(docs, toSpace, "/")
docs <- tm_map(docs, toSpace, "@")
docs <- tm_map(docs, toSpace, "\\|")
# Convert the text to lower case
docs <- tm_map(docs, content_transformer(tolower))
# Remove numbers
docs <- tm_map(docs, removeNumbers)
# Remove english common stopwords
docs <- tm_map(docs, removeWords, stopwords("english"))
# Remove your own stop word
# specify your stopwords as a character vector
docs <- tm_map(docs, removeWords, c("blabla1", "blabla2"))
# Remove punctuations
docs <- tm_map(docs, removePunctuation)
# Eliminate extra white spaces
docs <- tm_map(docs, stripWhitespace)
# Text stemming
# docs <- tm_map(docs, stemDocument)
dtm <- TermDocumentMatrix(docs)
m <- as.matrix(dtm)
v <- sort(rowSums(m),decreasing=TRUE)
d <- data.frame(word = names(v),freq=v)
head(d, 10)
set.seed(1234)
wordcloud(words = d$word, freq = d$freq, min.freq = 1,
max.words=200, random.order=FALSE, rot.per=0.35,
colors=brewer.pal(8, "Dark2"))
词云绘图结果每次布局都不一样哦,如下所示:
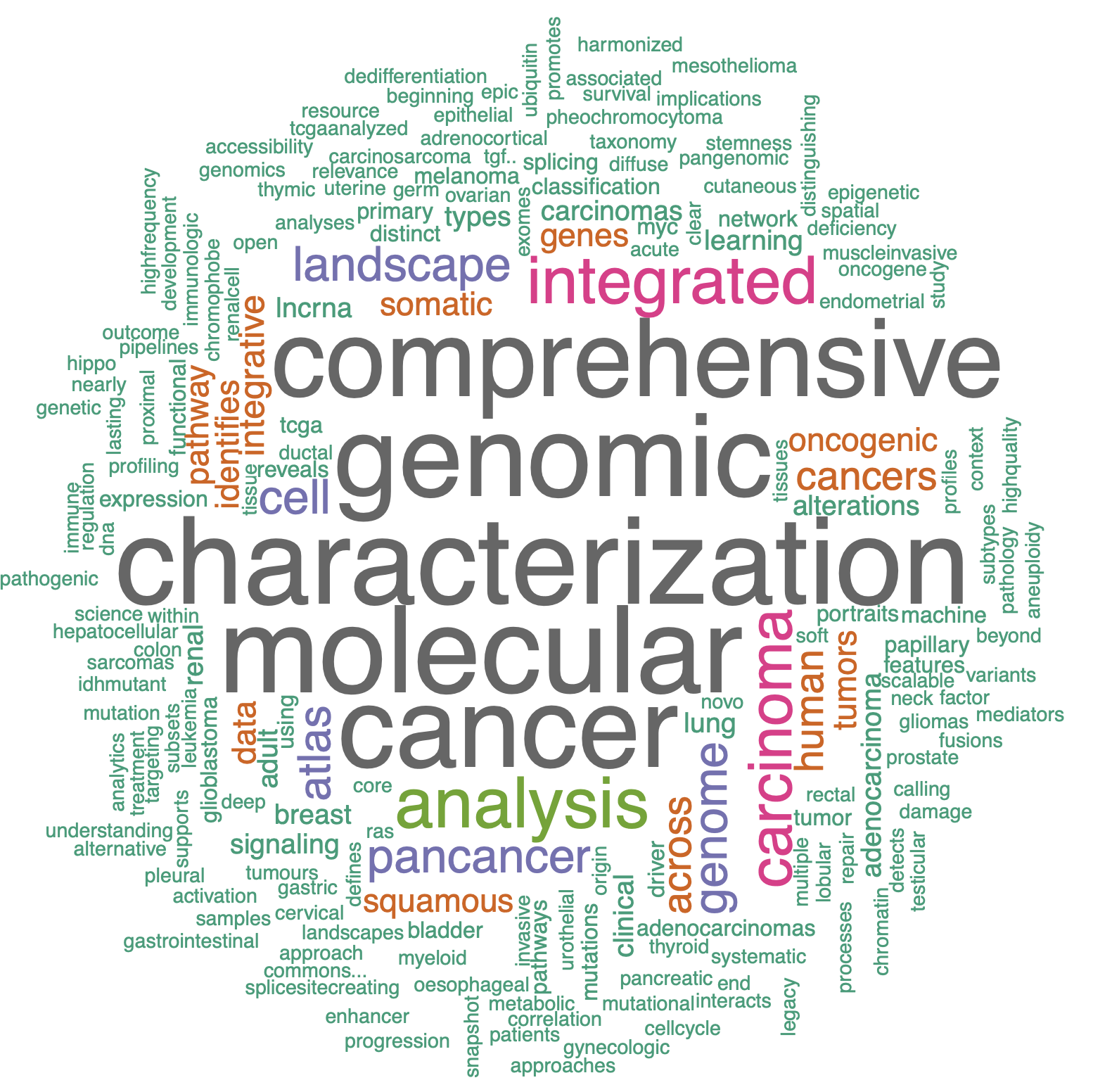
其实就是把词频给可视化了一下:
> head(d, 10)
word freq
1 characterization 25
2 molecular 25
3 genomic 24
4 cancer 23
5 comprehensive 22
6 analysis 13
7 integrated 12
8 carcinoma 11
9 cell 8
10 genome 8
出现次数很多的单词,在词云就显示大一点,仅此而已。
TCGA数据库其它系列教程
关于TCGA数据下载,我挑选了部分,写了6个数据下载系列教程:
- TCGA的28篇教程- 使用R语言的cgdsr包获取TCGA数据(cBioPortal)
- TCGA的28篇教程- 使用R语言的RTCGA包获取TCGA数据 (离线打包版本)
- TCGA的28篇教程-使用R语言的RTCGAToolbox包获取TCGA数据(FireBrowse portal)
- TCGA的28篇教程- 批量下载TCGA所有数据 ( UCSC的 XENA)
- TCGA的28篇教程-数据下载就到此为止吧
- TCGA的28篇教程-整理GDC下载的xml格式的临床资料
但是,建议你选择UCSC的xena数据库下载方式。如果你看视频,并不需要全盘接受,把握住重点。
也写了部分常见的TCGA数据库用法:
- TCGA的28篇教程-免疫全景图
- TCGA的28篇教程-指定癌症查看感兴趣基因的表达量
- TCGA的28篇教程-对TCGA数据库的任意癌症中任意基因做生存分析
- TCGA的28篇教程-风险因子关联图-一个价值1000但是迟到的答案
- TCGA的28篇教程-数据挖掘三板斧之ceRNA
- TCGA的28篇教程-所有癌症的突变全景图
- TCGA的28篇教程-早期泛癌研究
- TCGA的28篇教程-CNV全攻略
- TCGA的28篇教程-GTEx数据库-TCGA数据挖掘的好帮手
但是个人力量总归是有限的,我们生信技能树团队优秀R语言讲师《小洁》也学完了我的全套视频,在她自己的理解的基础上面,也给大家奉献了一套笔记: TCGA肿瘤数据库分析指南知识库马上面世
2008
Comprehensive genomic characterization defines human glioblastoma genes and core pathwaysExit Disclaimer
Nature. 2008;455(7216):1061-1068. doi:10.1038/nature07385
2011
Integrated genomic analyses of ovarian carcinomaExit Disclaimer
Nature. 2011;474(7353):609-615. doi:10.1038/nature10166
2012
Comprehensive molecular characterization of human colon and rectal cancerExit Disclaimer
Nature. 2012;487(7407):330-337. doi:10.1038/nature11252
Comprehensive molecular portraits of human breast tumoursExit Disclaimer
Nature. 2012;490(7418):61-70. doi:10.1038/nature11412
Comprehensive genomic characterization of squamous cell lung cancersExit Disclaimer
Nature. 2012;489(7417):519-525. doi:10.1038/nature11404
2013
Integrated genomic characterization of endometrial carcinomaExit Disclaimer
Nature. 2013;497(7447):67-73. doi:10.1038/nature12113
Genomic and epigenomic landscapes of adult de novo acute myeloid leukemiaExit Disclaimer
N Engl J Med. 2013;368(22):2059-2074. doi:10.1056/NEJMoa1301689
Comprehensive molecular characterization of clear cell renal cell carcinomaExit Disclaimer
Nature. 2013;499(7456):43-49. doi:10.1038/nature12222
The Cancer Genome Atlas Pan-Cancer analysis projectExit Disclaimer
Nat Genet. 2013;45(10):1113-1120. doi:10.1038/ng.2764
The somatic genomic landscape of glioblastoma01208-7)Exit Disclaimer
Cell. 2013;155(2):462-477. doi:10.1016/j.cell.2013.09.034
2014
Comprehensive molecular characterization of urothelial bladder carcinomaExit Disclaimer
Nature. 2014;507(7492):315-322. doi:10.1038/nature12965
Comprehensive molecular profiling of lung adenocarcinomaExit Disclaimer
Nature. 2014;511(7511):543-550. doi:10.1038/nature13385
Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin00876-9)Exit Disclaimer
Cell. 2014;158(4):929-944. doi:10.1016/j.cell.2014.06.049
The Somatic Genomic Landscape of Chromophobe Renal Cell Carcinoma00304-3)Exit Disclaimer
Cancer Cell. 2014;26(3):319-330. doi:10.1016/j.ccr.2014.07.014
Comprehensive molecular characterization of gastric adenocarcinomaExit Disclaimer
Nature. 2014;513(7517):202-209. doi:10.1038/nature13480
Integrated genomic characterization of papillary thyroid carcinoma01238-0)Exit Disclaimer
Cell. 2014;159(3):676-690. doi:10.1016/j.cell.2014.09.050
2015
Comprehensive genomic characterization of head and neck squamous cell carcinomasExit Disclaimer
Nature. 2015;517(7536):576-582. doi:10.1038/nature14129
Genomic Classification of Cutaneous Melanoma00634-0)Exit Disclaimer
Cell. 2015;161(7):1681-1696. doi:10.1016/j.cell.2015.05.044
Comprehensive, Integrative Genomic Analysis of Diffuse Lower-Grade GliomasExit Disclaimer
N Engl J Med. 2015;372(26):2481-2498. doi:10.1056/NEJMoa1402121
Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer01195-2)Exit Disclaimer
Cell. 2015;163(2):506-519. doi:10.1016/j.cell.2015.09.033
The Molecular Taxonomy of Primary Prostate Cancer01339-2)Exit Disclaimer
Cell. 2015;163(4):1011-1025. doi:10.1016/j.cell.2015.10.025
2016
Comprehensive Molecular Characterization of Papillary Renal-Cell CarcinomaExit Disclaimer
N Engl J Med. 2016;374(2):135-145. doi:10.1056/NEJMoa1505917
Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma30160-X)Exit Disclaimer
Cancer Cell. 2016;29(5):723-736. doi:10.1016/j.ccell.2016.04.002
Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomasExit Disclaimer
Nat Genet. 2016;48(6):607-616. doi:10.1038/ng.3564
2017
Integrated genomic characterization of oesophageal carcinomaExit Disclaimer
Nature. 2017;541(7636):169-175. doi:10.1038/nature20805
Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma30001-6)Exit Disclaimer
Cancer Cell. 2017;31(2):181-193. doi:10.1016/j.ccell.2017.01.001
Integrated Molecular Characterization of Uterine Carcinosarcoma30053-3)Exit Disclaimer
Cancer Cell. 2017;31(3):411-423. doi:10.1016/j.ccell.2017.02.010
Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles30214-0)Exit Disclaimer
Cell Rep. 2017;18(11):2780-2794. doi:10.1016/j.celrep.2017.02.033
Integrated genomic and molecular characterization of cervical cancerExit Disclaimer
Nature. 2017;543(7645):378-384. doi:10.1038/nature21386
Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma30639-6)Exit Disclaimer
Cell. 2017;169(7):1327-1341.e23. doi:10.1016/j.cell.2017.05.046
Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma30295-7)Exit Disclaimer
Cancer Cell. 2017;32(2):204-220.e15. doi:10.1016/j.ccell.2017.07.003
Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma30299-4)Exit Disclaimer
Cancer Cell. 2017;32(2):185-203.e13. doi:10.1016/j.ccell.2017.07.007
Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer31056-5)Exit Disclaimer
Cell. 2017;171(3):540-556.e25. doi:10.1016/j.cell.2017.09.007
Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas31203-5)Exit Disclaimer
Cell. 2017;171(4):950-965.e28. doi:10.1016/j.cell.2017.10.014
2018
The Integrated Genomic Landscape of Thymic Epithelial Tumors30003-5)Exit Disclaimer
Cancer Cell. 2018;33(2):244-258.e10. doi:10.1016/j.ccell.2018.01.003
Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas30097-8)Exit Disclaimer
Cell Syst. 2018;6(3):282-300.e2. doi:10.1016/j.cels.2018.03.003
Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines30096-6)Exit Disclaimer
Cell Syst. 2018;6(3):271-281.e7. doi:10.1016/j.cels.2018.03.002
Molecular Characterization and Clinical Relevance of Metabolic Expression Subtypes in Human Cancers30438-8)Exit Disclaimer
Cell Rep. 2018;23(1):255-269.e4. doi:10.1016/j.celrep.2018.03.077
Systematic Analysis of Splice-Site-Creating Mutations in Cancer30397-8)Exit Disclaimer
Cell Rep. 2018;23(1):270-281.e3. doi:10.1016/j.celrep.2018.03.052
Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types30152-9)Exit Disclaimer
Cell Rep. 2018;23(1):282-296.e4. doi:10.1016/j.celrep.2018.01.088
The Cancer Genome Atlas Comprehensive Molecular Characterization of Renal Cell Carcinoma30436-4)Exit Disclaimer
Cell Rep. 2018;23(1):313-326.e5. doi:10.1016/j.celrep.2018.03.075
Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context30425-X)Exit Disclaimer
Cell Rep. 2018;23(1):297-312.e12. doi:10.1016/j.celrep.2018.03.064
Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images30447-9)Exit Disclaimer
Cell Rep. 2018;23(1):181-193.e7. doi:10.1016/j.celrep.2018.03.086
Machine Learning Detects Pan-cancer Ras Pathway Activation in The Cancer Genome Atlas30389-9)Exit Disclaimer
Cell Rep. 2018;23(1):172-180.e3. doi:10.1016/j.celrep.2018.03.046
Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas30437-6)Exit Disclaimer
Cell Rep. 2018;23(1):239-254.e6. doi:10.1016/j.celrep.2018.03.076
Driver Fusions and Their Implications in the Development and Treatment of Human Cancers30395-4)Exit Disclaimer
Cell Rep. 2018;23(1):227-238.e3. doi:10.1016/j.celrep.2018.03.050
Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas30424-8)Exit Disclaimer
Cell Rep. 2018;23(1):194-212.e6. doi:10.1016/j.celrep.2018.03.063
Integrated Genomic Analysis of the Ubiquitin Pathway across Cancer Types30392-9)Exit Disclaimer
Cell Rep. 2018;23(1):213-226.e3. doi:10.1016/j.celrep.2018.03.047
SnapShot: TCGA-Analyzed Tumors30391-X)Exit Disclaimer
Cell. 2018;173(2):530. doi:10.1016/j.cell.2018.03.059
Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer30302-7)Exit Disclaimer
Cell. 2018;173(2):291-304.e6. doi:10.1016/j.cell.2018.03.022
The Cancer Genome Atlas: Creating Lasting Value beyond Its Data30374-X)Exit Disclaimer
Cell. 2018;173(2):283-285. doi:10.1016/j.cell.2018.03.042
Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation30358-1)Exit Disclaimer
Cell. 2018;173(2):338-354.e15. doi:10.1016/j.cell.2018.03.034
Oncogenic Signaling Pathways in The Cancer Genome Atlas30359-3)Exit Disclaimer
Cell. 2018;173(2):321-337.e10. doi:10.1016/j.cell.2018.03.035
Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics30313-1)Exit Disclaimer
Cell. 2018;173(2):305-320.e10. doi:10.1016/j.cell.2018.03.033
Comprehensive Characterization of Cancer Driver Genes and Mutations30237-X)Exit Disclaimer
Cell. 2018;173(2):371-385.e18. doi:10.1016/j.cell.2018.02.060
An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics30229-0)Exit Disclaimer
Cell. 2018;173(2):400-416.e11. doi:10.1016/j.cell.2018.02.052
Pathogenic Germline Variants in 10,389 Adult Cancers30363-5)Exit Disclaimer
Cell. 2018;173(2):355-370.e14. doi:10.1016/j.cell.2018.03.039
A Pan-Cancer Analysis of Enhancer Expression in Nearly 9000 Patient Samples30307-6)Exit Disclaimer
Cell. 2018;173(2):386-399.e12. doi:10.1016/j.cell.2018.03.027
Genomic and Functional Approaches to Understanding Cancer Aneuploidy30111-9)Exit Disclaimer
Cancer Cell. 2018;33(4):676-689.e3. doi:10.1016/j.ccell.2018.03.007
A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers30119-3)Exit Disclaimer
Cancer Cell. 2018;33(4):690-705.e9. doi:10.1016/j.ccell.2018.03.014
Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas30114-4)Exit Disclaimer
Cancer Cell. 2018;33(4):721-735.e8. doi:10.1016/j.ccell.2018.03.010
lncRNA Epigenetic Landscape Analysis Identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and Promotes Cell-Cycle Progression in Cancer30110-7)Exit Disclaimer
Cancer Cell. 2018;33(4):706-720.e9. doi:10.1016/j.ccell.2018.03.006
The Immune Landscape of Cancer30121-3)Exit Disclaimer
Immunity. 2018;48(4):812-830.e14. doi:10.1016/j.immuni.2018.03.023
Integrated Molecular Characterization of Testicular Germ Cell Tumors30785-X)Exit Disclaimer
Cell Rep. 2018;23(11):3392-3406. doi:10.1016/j.celrep.2018.05.039
Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients30306-4)Exit Disclaimer
Cancer Cell. 2018;34(2):211-224.e6. doi:10.1016/j.ccell.2018.07.001
A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily30357-0)Exit Disclaimer
Cell Systems. 2018;7(4);422-437.e7. doi: 10.1016/j.cels.2018.08.010
Integrative Molecular Characterization of Malignant Pleural MesotheliomaExit Disclaimer
Cancer Discovery. 2018;8(12):1548-1565. doi: 10.1158/2159-8290.CD-18-0804
The chromatin accessibility landscape of primary human cancersExit Disclaimer
Science. 2018;362(6413). pii: eaav1898. doi: 10.1126/science.aav1898
Comprehensive Molecular Characterization of the Hippo Signaling Pathway in Cancer31564-X)Exit Disclaimer
Cell Reports. 2018;25(5):1304-1317.e5. doi: 10.1016/j.celrep.2018.10.001
2019
Before and After: Comparison of Legacy and Harmonized TCGA Genomic Data Commons’ Data30201-7)Exit Disclaimer
Cell Systems. 2019;9(1):24-34.e10. doi: 10.1016/j.cels.2019.06.006
2020
Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer30211-7)Exit Disclaimer
Cancer Cell. 2020;37(5):639-654.e6. doi: 10.1016/j.ccell.2020.04.012
文末友情推荐
要想真正入门生物信息学建议务必购买全套书籍,一点一滴攻克计算机基础知识,书单在:什么,生信入门全套书籍仅需160 。
如果大家没有时间自行慢慢摸索着学习,可以考虑我们生信技能树官方举办的学习班:
- 数据挖掘学习班第7期(线上直播3周,马拉松式陪伴,带你入门),原价4800的数据挖掘全套课程, 疫情期间半价即可抢购。
- 生信爆款入门-第9期(线上直播4周,马拉松式陪伴,带你入门),原价9600的生信入门全套课程,疫情期间3.3折即可抢购。
如果你课题涉及到转录组,欢迎添加一对一客服:详见:你还在花三五万做一个单细胞转录组吗?
号外:生信技能树知识整理实习生招募,长期招募,也可以简单参与软件测评笔记撰写,开启你的分享人生!另外,:绝大部分生信技能树粉丝都没有机会加我微信,已经多次满了5000好友,所以我开通了一个微信好友,前100名添加我,仅需150元即可,3折优惠期机会不容错过哈。我的微信小号二维码在:0元,10小时教学视频直播《跟着百度李彦宏学习肿瘤基因组测序数据分析》