粉丝来信求助,他感兴趣的一个数据集的芯片平台是:GE Healthcare/Amersham Biosciences CodeLink Human Whole Genome Bioarray,链接是:GPL2895
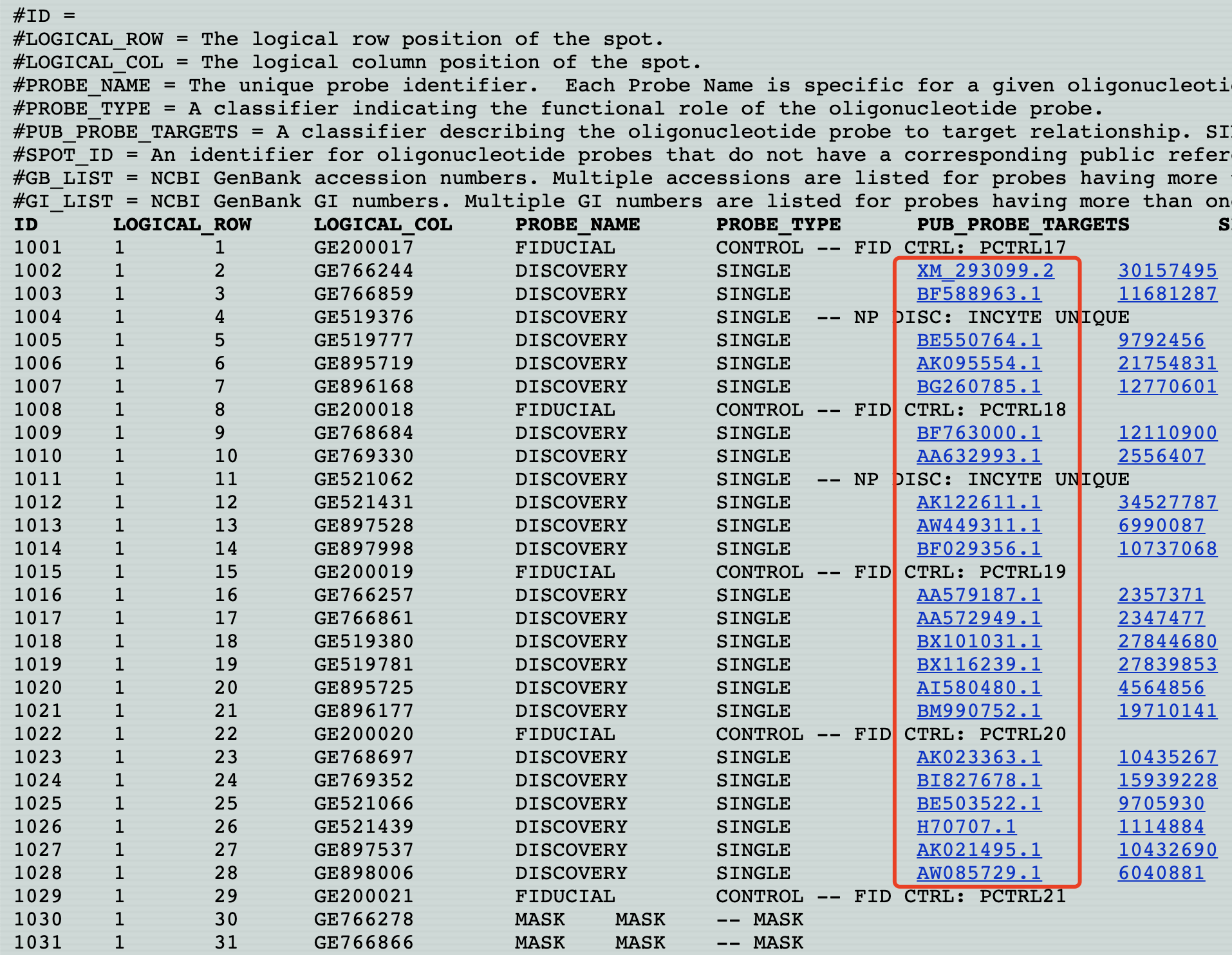
提供的芯片注释信息是 NCBI GenBank accession numbers ,如下所示:
#GB_LIST = NCBI GenBank accession numbers. Multiple accessions are listed for probes having more than one target transcript (the first accession in the list is the primary accession assigned to the target sequence)
#GI_LIST = NCBI GenBank GI numbers. Multiple GI numbers are listed for probes having more than one target transcript (the first GI number in the list is the primary GI assigned to the target sequence)
他已经很敏锐的发现了这个 NCBI GenBank accession numbers 就是基因的信息:
随便点击一个进去:https://www.ncbi.nlm.nih.gov/nuccore/AW027747.1
也确实可以看到它的详情:
/organism="Homo sapiens"
/mol_type="mRNA"
/db_xref="taxon:9606"
/clone="IMAGE:2535419"
/clone_lib="SAMN00156302 Soares_thymus_NHFTh"
/dev_stage="fetal"
/lab_host="DH10B (phage-resistant)"
/note="Organ: thymus, pooled; Vector: pT7T3D-PacI; Site_1:
Not I; Site_2: Eco RI; 1st strand cDNA was primed with a
Not I - oligo(dT) primer [5'
TGTTACCAATCTGAAGTGGGAGCGGCCGCAACGTTTTTTTTTTTTTTTTTT 3'],
double-stranded cDNA was ligated to Eco RI adaptors
(Pharmacia), digested with Not I and cloned into the Not I
and Eco RI sites of the modified pT7T3 vector. Library
went through one round of normalization. Library
constructed by Bento Soares and M. Fatima Bonaldo."
但是不可能这样的手动一个个去查它的信息,需要使用R代码进行转换。
查看org.Hs.eg.db包的近80万个GenBank的索引号
library(org.Hs.eg.db)
keytypes(org.Hs.eg.db)
tmp=toTable(org.Hs.egACCNUM)
head(tmp)
可以看到一个基因对应多个GenBank的索引号,因为2万个基因却有80万个GenBank的索引号
> head(tmp)
gene_id accession
1 1 AA484435
2 1 AAH35719
3 1 AAL07469
4 1 AB073611
5 1 ACJ13639
6 1 AF414429
这个包有80万个GenBank的索引号,但是未必是最全面或者说最新的信息,不过,目前是最方便的解决方案,所以这里我推荐给大家。
然后读取GPL2895的5万注释信息
自己在 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?view=data&acc=GPL2895&id=20430&db=GeoDb_blob09 里面复制粘贴成为 GPL2895.txt 这个文本文件。
代码如下:
a=read.table('GPL2895.txt',sep = '\t',header = T)
# 下面这个错误的居然是1.7G的压缩文本
# a=read.table('~/Desktop/GPL2895_family.soft.gz',sep = '\t',header = T)
head(a)
a=a[nchar(a$GB_LIST)>1,]
a=a[,c(1,8)]
head(a)
library(stringr)
a$GB_LIST=str_split(a$GB_LIST,'[.]',simplify = T)[,1]
head(a)
这样GPL2895芯片的探针就对应到了 GenBank的索引号
> head(a)
ID GB_LIST
2 1002 XM_293099
3 1003 BF588963
5 1005 BE550764
6 1006 AK095554
7 1007 BG260785
9 1009 BF763000
最后关联一下
b=merge(a,tmp,by.x='GB_LIST',by.y='accession')
head(b)
d=toTable(org.Hs.egSYMBOL)
f=merge(b,d,by='gene_id')
head(f)
得到的结果如下:
> head(f)
gene_id GB_LIST ID symbol
1 1 W25099 338005 A1BG
2 10 NM_000015 118081 NAT2
3 100 NM_000022 494110 ADA
4 1000 NM_001792 490046 CDH2
5 10000 U79271 318033 AKT3
6 10000 NM_181690 457002 AKT3
可以看到,芯片的探针的各种ID都关联起来啦。
是不是超级简单啊!