我们做肿瘤研究的单细胞数据,一般来说初步定义细胞亚群, 第一次分群是通用规则,按照 :
- immune (CD45+,PTPRC),
- epithelial/cancer (EpCAM+,EPCAM),
- stromal (CD10+,MME,fibo or CD31+,PECAM1,endo)
然后绝大部分都是抓住免疫细胞亚群进行细分,包括淋巴系(T,B,NK细胞)和髓系(单核,树突,巨噬,粒细胞)的两大类作为第二次细分亚群。第三次细分亚群就会深入到T淋巴细胞的CD8和CD4细分,这个时候很多人就很难搞起来各个细分亚群的标记基因了,最近( 06 May 2021)出来了一个文章:《A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer》,链接是:https://www.nature.com/articles/s41591-021-01323-8
这方面就做的很细致,如下所示的基因:

但是这么多基因名字,如果你也想把它用起来,得一个个手敲。这当然是不符合咱们现代人的习惯啦,我们善假于物,所以我随手找了一个网页工具:http://www.ocrmaker.com/
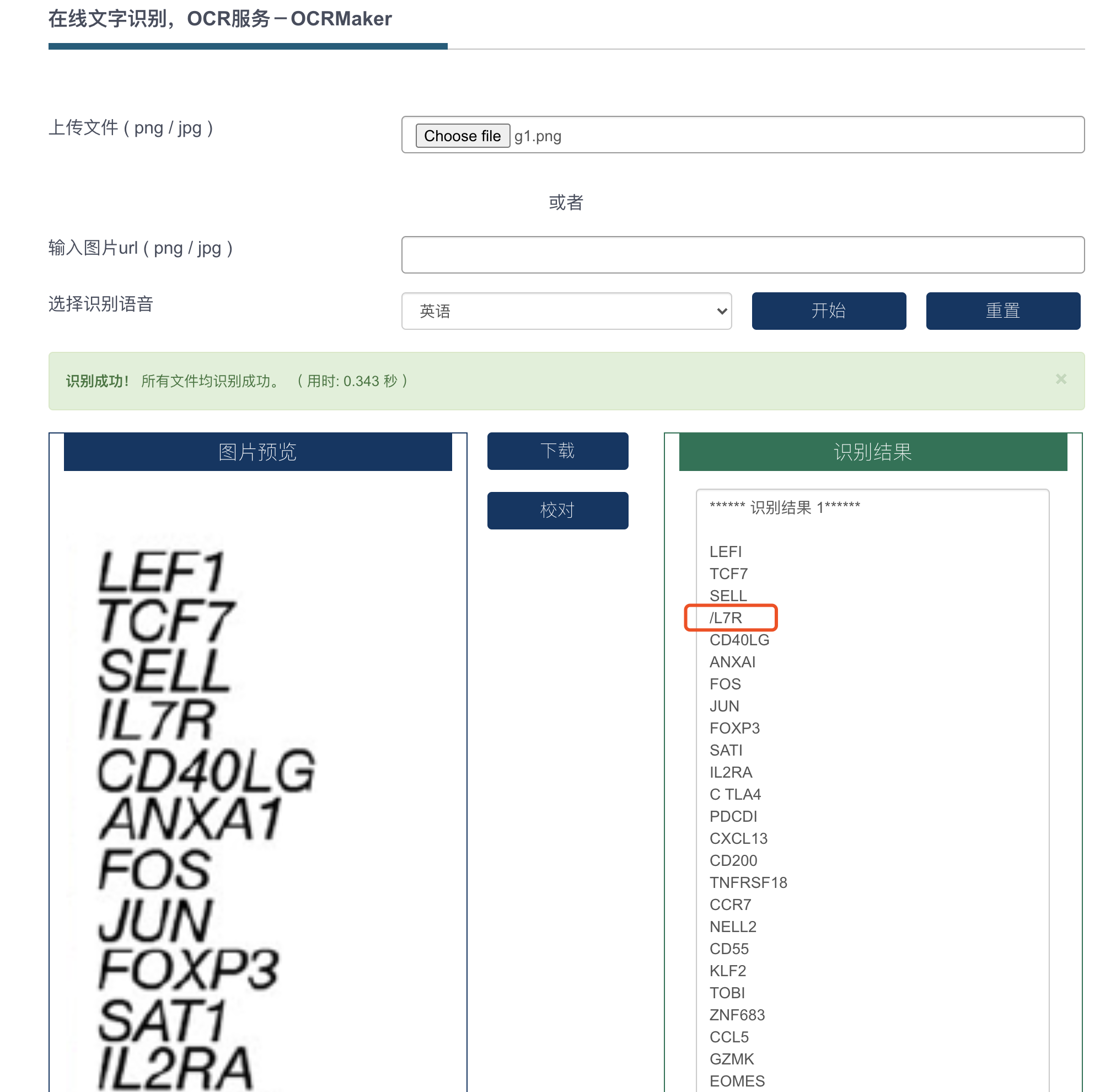
结果可以接受!其实主要是1和I这两个很难区分,修改回来即可:
The following requested variables were not found (10 out of 13 shown):
LEFI, ANXAI, SATI, PDCDI, TOBI, FCGRGA, KLRCI, KLRDI, TRDVI, MK167
需要一些手动修改,最后绘图代码如下:
rm(list=ls())
options(stringsAsFactors = F)
library(Seurat)
load(file = 'main_sce_recluster.Rdata')
marker_genes= c("LEF1","TCF7","SELL","IL7R","CD40LG","ANXA1","FOS",
"JUN","FOXP3","SAT1","IL2RA","CTLA4","PDCD1","CXCL13","CD200",
"TNFRSF18","CCR7","NELL2","CD55","KLF2","TOB1","ZNF683","CCL5",
"GZMK","EOMES","ITM2C","CX3CR1","GNLY","GZMH","GZMB","LAG3","CCL4L2",
"FCGR3A","FGFBP2","TYROBP","AREG","XCL1","KLRC1","TRDV2","TRGV9","MTRNR2L8",
"KLRD1","TRDV1","KLRC3",
"CTSW","CD7","MKI67","STMN1","TUBA1B","HIST1H4C" )
p <- DotPlot(sce, features = marker_genes,
assay='RNA' ,group.by = 'cellSubType' ) + coord_flip() +
theme(axis.text.x = element_text(angle = 90))
p
ggsave('check_g1_markers_by_cellSubType.pdf')
marker_genes =c("CD3D","CD4","CD8A","CCR7","LEF1","SELL","TCF7","GNLY","IFNG","NKG7","PRF1",
"GZMA","GZMB","GZMH","GZMK","HAVCR2","LAG3","PDCD1","CTLA4","TIGIT","BTLA","KLRC1",
"ANXA1","ANKRD28","CD69","CD40LG","ZNF683","FOXP3","IL2RA","IKZF2","NCR1","NCAM1",
"TYROBP","KLRD1","KLRF1","KLRB1","CX3CR1","FCGR3A",
"XCL1","XCL2","TRDV2","TRGV9","TRGC2","MKI67","TOP2A")
p <- DotPlot(sce, features = marker_genes,
assay='RNA' ,group.by = 'cellSubType' ) + coord_flip() +
theme(axis.text.x = element_text(angle = 90))
p
ggsave('check_g2_markers_by_cellSubType.pdf')
可以看到效果还不错!

如果你也有T淋巴细胞的CD8和CD4细分需求,这些基因拿去使用吧!
工具并不是万能的
比如当我想上传的图片里面的基因名字有一些重叠的时候 :
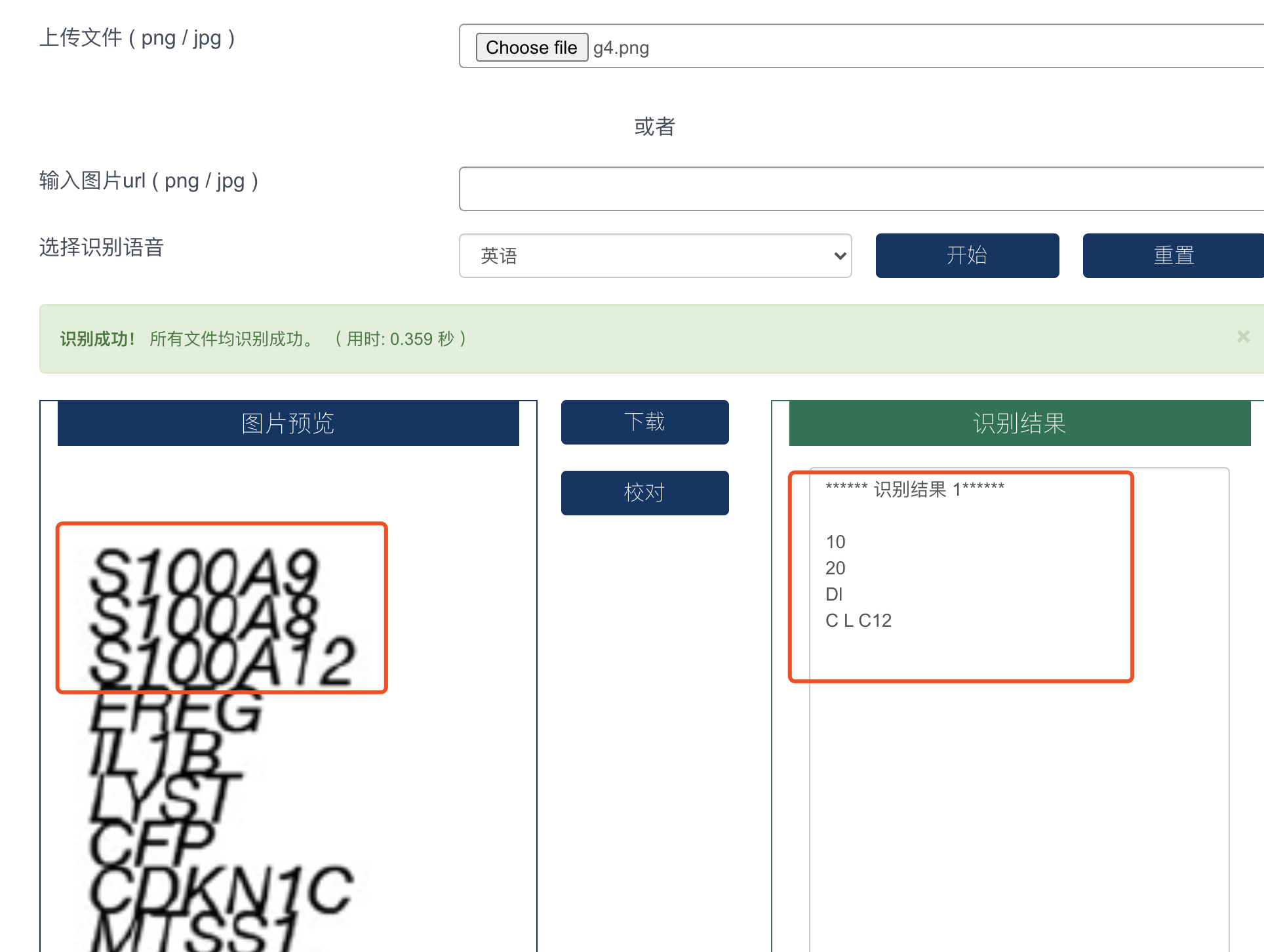
这个时候肉眼其实是可以区分的, 所以你还可以退而求其次,选择比较廉价的本科生帮你手敲这些基因名字哦!
学徒作业
我有一个CNS图表复现专辑,里面的CD8和CD4就是可以细分,大家完成降维聚类分群后,使用我们这个教程里面的基因名字进行细分亚群,并且合理的展现!
- CNS图表复现01—读入csv文件的表达矩阵构建Seurat对象
- CNS图表复现02—Seurat标准流程之聚类分群
- CNS图表复现03—单细胞区分免疫细胞和肿瘤细胞
- CNS图表复现04—单细胞聚类分群的resolution参数问题
- CNS图表复现05—免疫细胞亚群再分类
- CNS图表复现06—根据CellMarker网站进行人工校验免疫细胞亚群
- CNS图表复现07—原来这篇文章有两个单细胞表达矩阵
- CNS图表复现08—肿瘤单细胞数据第一次分群通用规则
- CNS图表复现09—上皮细胞可以区分为恶性与否
- CNS图表复现10—表达矩阵是如何得到的
- CNS图表复现11—RNA-seq数据可不只是表达量矩阵结果
- CNS图表复现12—检查原文的细胞亚群的标记基因
- CNS图表复现13—使用inferCNV来区分肿瘤细胞的恶性与否
- CNS图表复现14—检查文献的inferCNV流程
- CNS图表复现15—inferCNV流程输入数据差异大揭秘
- CNS图表复现16—inferCNV结果解读及利用
- CNS图表复现17—inferCNV结果解读及利用之进阶