以前我们做了一个投票:可视化单细胞亚群的标记基因的5个方法,下面的5个基础函数相信大家都是已经烂熟于心了:
- VlnPlot(pbmc, features = c(“MS4A1”, “CD79A”))
- FeaturePlot(pbmc, features = c(“MS4A1”, “CD79A”))
- RidgePlot(pbmc, features = c(“MS4A1”, “CD79A”), ncol = 1)
- DotPlot(pbmc, features = unique(features)) + RotatedAxis()
- DoHeatmap(subset(pbmc, downsample = 100), features = features, size = 3)
但是架不住一些变态老板或者说甲方的无止境修改需求,看到一个好的图表就让你去复现,真的很坑嗲啊!明明都是一个意思,就是某个亚群的某个基因特异性表达而已,非要玩出花!
比如2020的文章:《The Molecular Anatomy of Mouse Skin during Hair Growth and Rest》就另辟蹊径,出来了如下的可视化方法:
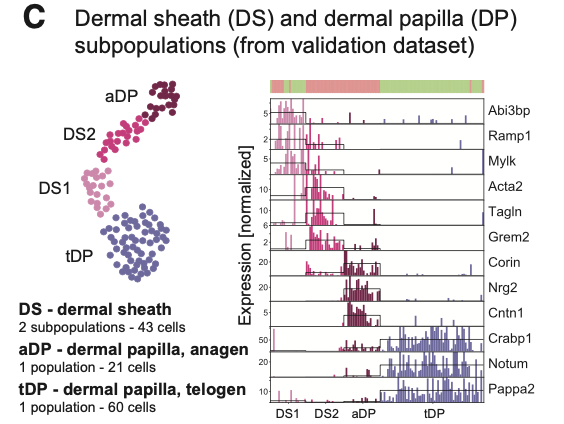
图例是:(C) UMAP of dermal papilla (DP) and dermal sheath (DS) cells from the validation dataset (left) and marker gene expression associated with each subpopulation (right). Black lines, expression average for each subpopulation. Top panel, hair cycle stage of each cell (green, telogen; pink, anagen).
很明显它并不是 Seurat 流程内置的图表,如果你一定要使用R语言绘制它也不是不可以,但是如果你仔细看文章,就会发现它同时也提到了 Scanpy 这个基于python编程语言的包。
值得一提的是我在文章:《A molecular atlas of cell types and zonation in the brain vasculature》也看到了类似的展示方式:
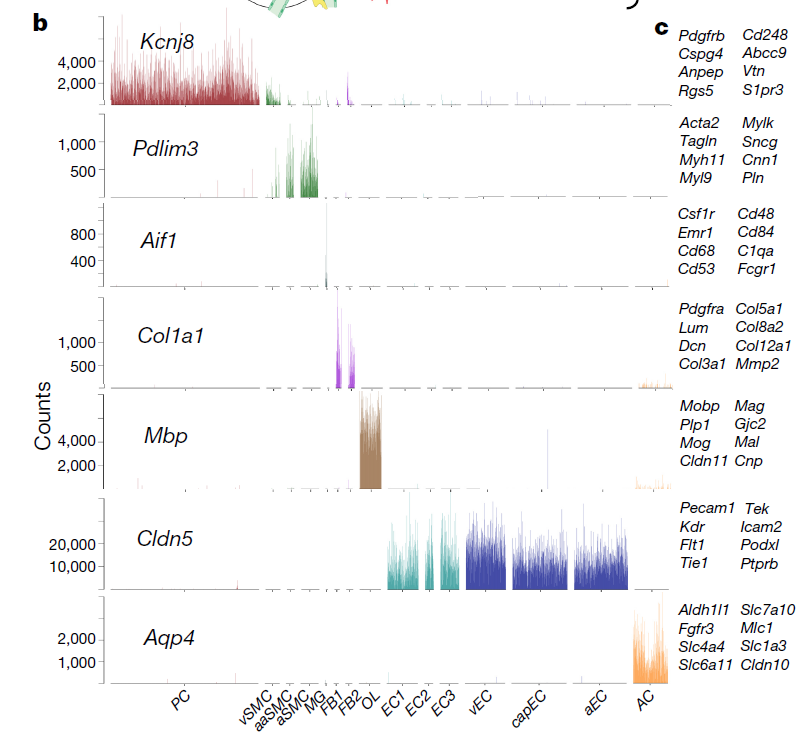
同样的,也是 Scanpy 这个基于python编程语言的包。
那么现在问题来了,你更倾向于哪个包呢?