看到了朋友圈转发一个最新的(November 2021)发表在nature杂志的研究成果,日本RIKEN的科研团队首次发现,B细胞竟会分泌一种神经递质——γ-氨基丁酸(GABA),进而抑制细胞毒性T细胞功能、促进肿瘤相关巨噬细胞分化,抑制抗肿瘤免疫!除去B细胞之后,小鼠体内的抗肿瘤免疫也变强了。这个研究也是有转录组测序数据的,在GSE169543 and GSE183246.。参考资料:
[1] https://www.nature.com/articles/s41586-021-04082-1
[2] https://www.nature.com/articles/s41423-018-0027-x
[3] https://www.nature.com/articles/d41586-021-02953-1
首先拿到GABA相关基因
在日本的kegg数据库可以查询:https://www.genome.jp/kegg/pathway.html
这个 GABAergic synapse 在 https://www.genome.jp/entry/pathway+hsa04727 ,进入可以查询其基因
54407 SLC38A2; solute carrier family 38 member 2 [KO:K14207]
27165 GLS2; glutaminase 2 [KO:K01425] [EC:3.5.1.2]
2744 GLS; glutaminase [KO:K01425] [EC:3.5.1.2]
2571 GAD1; glutamate decarboxylase 1 [KO:K01580] [EC:4.1.1.15]
2572 GAD2; glutamate decarboxylase 2 [KO:K01580] [EC:4.1.1.15]
140679 SLC32A1; solute carrier family 32 member 1 [KO:K15015]
------------ 此处省略几十个基因 ------------
92745 SLC38A5; solute carrier family 38 member 5 [KO:K14992]
在pbmc3k查询该基因列表
如果你不熟悉这个 pbmc3k 单细胞数据集,自己去看前面的笔记哈,反正是下面的代码即可下载:
install.packages('devtools')
devtools::install_github('satijalab/seurat-data')
library(SeuratData) #加载seurat数据集
getOption('timeout')
options(timeout=10000)
InstallData("pbmc3k")
data("pbmc3k")
既然咱们的电脑里面有了 pbmc3k 单细胞数据集,接下来就可以查看我们的 GABA相关基因在这个数据集的表现情况啦:
library(stringr)
genes_to_check = c("SLC38A1","SLC38A2","GLS2","GLS","GAD1","GAD2","SLC32A1","ABAT","GABRA1","GABRA2","GABRA3","GABRA4","GABRA5","GABRA6","GABRB1","GABRB3","GABRB2","GABRG1","GABRG2","GABRG3","GABRD","GABRE","GABRQ","GABRP","PRKACA","PRKACB","PRKACG","SRC","PRKCA","PRKCB","PRKCG","HAP1","GABARAP","GABARAPL1","GABARAPL2","NSF","TRAK2","PLCL1","GPHN","SLC12A5","GABRR1","GABRR2","GABRR3","CACNA1A","CACNA1B","CACNA1C","CACNA1D","CACNA1F","CACNA1S","GABBR1","GABBR2","GNAI1","GNAI3","GNAI2","GNAO1","GNB1","GNB2","GNB3","GNB4","GNB5","GNG2","GNG3","GNG4","GNG5","GNG7","GNG8","GNG10","GNG11","GNG12","GNG13","GNGT1","GNGT2","ADCY1","ADCY2","ADCY3","ADCY4","ADCY5","ADCY6","ADCY7","ADCY8","ADCY9","KCNJ6","SLC6A1","SLC6A13","SLC6A11","SLC6A12","GLUL","SLC38A3","SLC38A5")
# 该基因名称(大小写转换)
genes_to_check=str_to_upper(genes_to_check)
genes_to_check
library(Seurat)
library(ggplot2)
library(SeuratData)
data("pbmc3k")
table(Idents(pbmc3k.final))
p_all_markers <- DotPlot(pbmc3k.final,
features = genes_to_check,
assay='RNA' ) + coord_flip()+theme(axis.text.x = element_text(angle = 45,
vjust = 0.5, hjust=0.5))
p_all_markers
ggsave(plot=p_all_markers,
filename="GABA_genes_in_pbmc3k.pdf",width = 6,height = 10)
library(cowplot)
p_umap=DimPlot(pbmc3k.final,label = T,repel = T)
p1 <- plot_grid(p_all_markers,p_umap,rel_widths = c(1.5,1))
p1
ggsave(p1,filename = 'GABA_genes_in_pbmc3k_and_umap.pdf',
units = "cm",width = 35,height = 18)
# The following requested variables were not found (10 out of 46 shown):
# GAD1, GAD2, SLC32A1, GABRA1, GABRA2, GABRA3, GABRA4, GABRA5, GABRA6, GABRB1
可以看到,这个 pbmc3k 单细胞数据集,其实很多基因是缺少的。
可以看到B细胞并没有特殊之处哦:

当然了,需要跟降维聚类分群结果对应,如下所示:
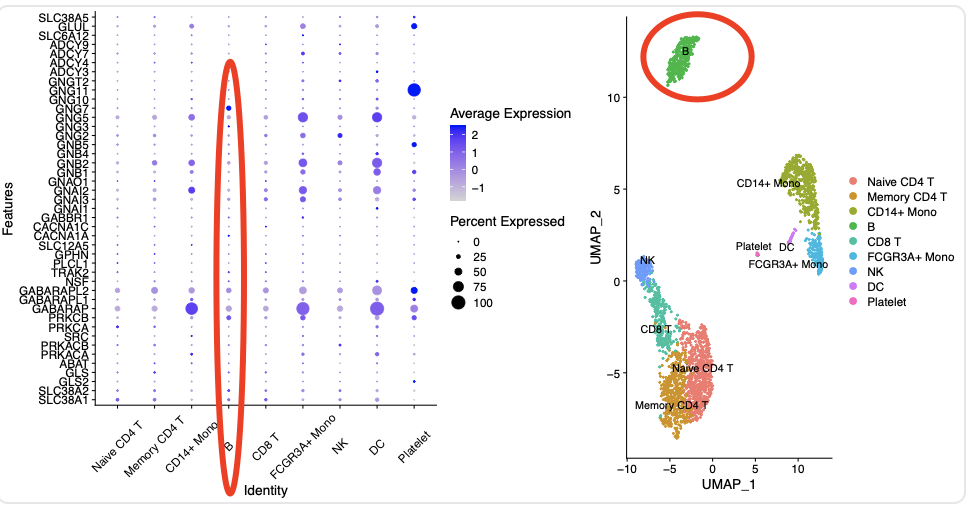
从全局表达量来说,B细胞其实是非常特殊了,可惜,它的 GABA相关基因 并没有特殊之处
在AML疾病单细胞数据集查询该基因列表
因为涉及到了AML疾病单细胞数据集处理,是冗长代码,这里略,单细胞转录组数据分析的标准降维聚类分群,并且进行生物学注释后的结果。可以参考前面的例子:人人都能学会的单细胞聚类分群注释 ,我们演示了第一层次的分群。
如果你对单细胞数据分析还没有基础认知,可以看基础10讲:
- 01. 上游分析流程
- 02.课题多少个样品,测序数据量如何
- 03. 过滤不合格细胞和基因(数据质控很重要)
- 04. 过滤线粒体核糖体基因
- 05. 去除细胞效应和基因效应
- 06.单细胞转录组数据的降维聚类分群
- 07.单细胞转录组数据处理之细胞亚群注释
- 08.把拿到的亚群进行更细致的分群
- 09.单细胞转录组数据处理之细胞亚群比例比较
我们直接看效果:
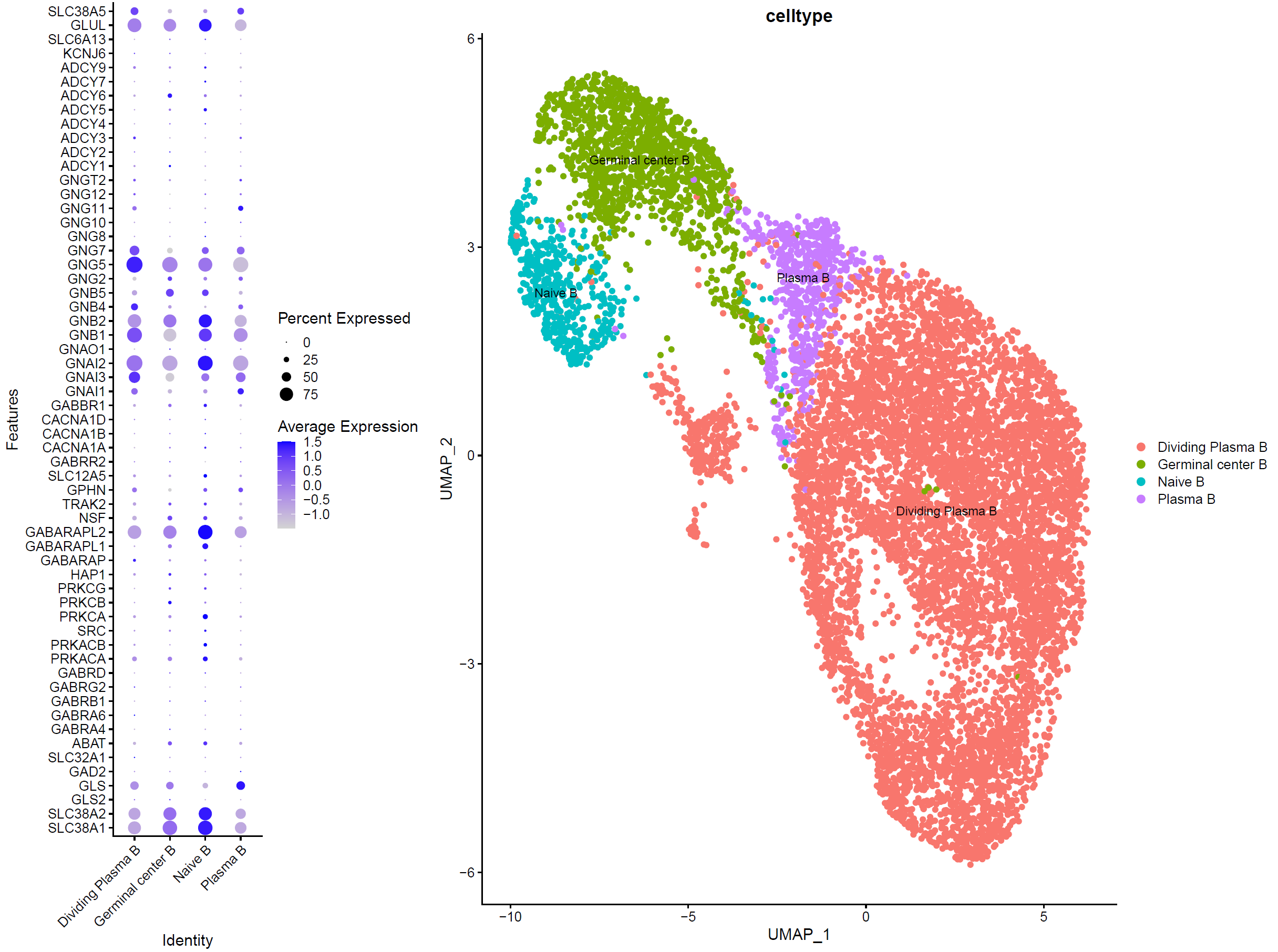
可以看到, b细胞各个亚群都或多或少表达GABA相关基因,那么这样的结论值得发表在 nature杂志 吗?
甚至完全没有单细胞高级分析啊 ,比如转录因子分析 ,主要是对计算机资源消耗会比较大,我也多次分享过细节教程: