在朋友圈看到了一个单细胞文献快讯:杜克-新加坡国立大学医学院和新加坡科学技术局基因组研究所等机构的研究人员在 Cancer Discovery 期刊发表了题为:《Single-cell atlas of lineage states, tumor microenvironment and subtypespecific expression programs in gastric cancer》 的研究论文。 关于单细胞数据来源的介绍是:We generated a comprehensive single-cell atlas of GC comprising 31 primary gastric tumor samples representing clinical stages I-IV and histological subtypes. 更加具体的样品信息在 Supplementary Table 1 , 我们这里就不展开介绍该研究做了什么,直接单刀直入,看其单细胞数据情况!
我看了看,这个数据集是公开的,链接是:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE183904 ,可以看到是:
GSM5573466 sample1: Primary Gastric Tissue (Normal)
GSM5573467 sample2: Primary Gastric Tissue (Tumor)
GSM5573468 sample3: Primary Gastric Tissue (Tumor)
GSM5573469 sample4: Primary Gastric Tissue (Normal)
GSM5573470 sample5: Primary Gastric Tissue (Tumor)
GSM5573471 sample6: Primary Gastric Tissue (Normal)
GSM5573472 sample7: Primary Gastric Tissue (Tumor)
GSM5573473 sample8: Primary Gastric Tissue (Tumor)
GSM5573474 sample9: Primary Gastric Tissue (Normal)
GSM5573475 sample10: Primary Gastric Tissue (Tumor)
GSM5573476 sample11: Primary Gastric Tissue (Normal)
GSM5573477 sample12: Primary Gastric Tissue (Tumor)
GSM5573478 sample13: Primary Gastric Tissue (Tumor)
GSM5573479 sample14: Primary Gastric Tissue (Tumor)
GSM5573480 sample15: Primary Gastric Tissue (Tumor)
GSM5573481 sample16: Primary Gastric Tissue (Tumor)
GSM5573482 sample17: Primary Gastric Tissue (Tumor)
GSM5573483 sample18: Primary Gastric Tissue (Tumor)
GSM5573484 sample19: Peritonium tissue (Tumor)
GSM5573485 sample20: Peritonium tissue (Tumor)
GSM5573486 sample21: Primary Gastric Tissue (Normal)
GSM5573487 sample22: Primary Gastric Tissue (Tumor)
GSM5573488 sample23: Primary Gastric Tissue (Normal)
GSM5573489 sample24: Primary Gastric Tissue (Tumor)
GSM5573490 sample25: Primary Gastric Tissue (Normal)
GSM5573491 sample26: Primary Gastric Tissue (Tumor)
GSM5573492 sample27: Primary Gastric Tissue (Tumor)
GSM5573493 sample28: Primary Gastric Tissue (Tumor)
GSM5573494 sample29: Primary Gastric Tissue (Tumor)
GSM5573495 sample30: Primary Gastric Tissue (Tumor)
GSM5573496 sample31: Primary Gastric Tissue (Normal)
GSM5573497 sample32: Primary Gastric Tissue (Tumor)
GSM5573498 sample33: Primary Gastric Tissue (Tumor)
GSM5573499 sample34: Primary Gastric Tissue (Tumor)
GSM5573500 sample35: Primary Gastric Tissue (Normal)
GSM5573501 sample36: Primary Gastric Tissue (Tumor)
GSM5573502 sample37: Peritonium tissue (Normal)
GSM5573503 sample38: Peritonium tissue (Tumor)
GSM5573504 sample39: Primary Gastric Tissue (Tumor)
GSM5573505 sample40: Primary Gastric Tissue (Tumor)
其分群基本上跟我们提到了的肿瘤单细胞分群规则类似的,首先是按照如下所示的标记基因进行第一次分群 :
- immune (CD45+,PTPRC),
- epithelial/cancer (EpCAM+,EPCAM),
- stromal (CD10+,MME,fibo or CD31+,PECAM1,endo)
然后每个亚群进行第二层次细分亚群,甚至第三层次,第四次分群,结构清晰明了。
文章也是如此,第一层次是:
- immune cell populations dominated the cell-states (21 of 34 states).
- The “Epithelial” meta-cluster (7 cell-states; CDH1 positive) contained three distinct sub-lineages;
- The “Stromal meta-cluster” (6 cell-states; FN1 positive)
然后各自细分亚群,首先是免疫细胞:
- (1) a “Myeloid meta-cluster” (5 cell-states),
- (2) a “Lymphoid meta- cluster” (11 cell-states),
- (3) “Plasma meta-cluster” (5 cell-states),
以及肿瘤相关的基质细胞:
- pericytes (STF2; defined by RGS5 and NOTCH3),
- fibroblasts (STF1 and STF3, defined by LUM and DCN),
- PLVAP positive endothelial cell subclusters.
那我们现在就带领大家快速检查迄今为止时间上最大的胃癌单细胞队列数据质量,首先下载这个GSE183904_RAW.tar 329.2 Mb文件,并且解压,如下所示:
6.0M 9 11 00:19 GSM5573466_sample1.csv.gz
6.7M 9 11 00:19 GSM5573467_sample2.csv.gz
9.1M 9 11 00:19 GSM5573468_sample3.csv.gz
4.9M 9 11 00:19 GSM5573469_sample4.csv.gz
8.8M 9 11 00:19 GSM5573470_sample5.csv.gz
4.9M 9 11 00:19 GSM5573471_sample6.csv.gz
5.2M 9 11 00:19 GSM5573472_sample7.csv.gz
15M 9 11 00:19 GSM5573473_sample8.csv.gz
6.3M 9 11 00:19 GSM5573474_sample9.csv.gz
5.4M 9 11 00:19 GSM5573475_sample10.csv.gz
2.4M 9 11 00:19 GSM5573476_sample11.csv.gz
1.8M 9 11 00:19 GSM5573477_sample12.csv.gz
3.0M 9 11 00:19 GSM5573478_sample13.csv.gz
13M 9 11 00:20 GSM5573479_sample14.csv.gz
28M 9 11 00:20 GSM5573480_sample15.csv.gz
4.5M 9 11 00:20 GSM5573481_sample16.csv.gz
17M 9 11 00:20 GSM5573482_sample17.csv.gz
11M 9 11 00:20 GSM5573483_sample18.csv.gz
4.1M 9 11 00:20 GSM5573484_sample19.csv.gz
10M 9 11 00:20 GSM5573485_sample20.csv.gz
4.7M 9 11 00:20 GSM5573486_sample21.csv.gz
1.0M 9 11 00:20 GSM5573487_sample22.csv.gz
7.4M 9 11 00:20 GSM5573488_sample23.csv.gz
4.9M 9 11 00:21 GSM5573489_sample24.csv.gz
9.6M 9 11 00:21 GSM5573490_sample25.csv.gz
5.5M 9 11 00:21 GSM5573491_sample26.csv.gz
7.0M 9 11 00:21 GSM5573492_sample27.csv.gz
13M 9 11 00:21 GSM5573493_sample28.csv.gz
9.0M 9 11 00:21 GSM5573494_sample29.csv.gz
6.2M 9 11 00:21 GSM5573495_sample30.csv.gz
4.2M 9 11 00:21 GSM5573496_sample31.csv.gz
5.1M 9 11 00:21 GSM5573497_sample32.csv.gz
13M 9 11 00:21 GSM5573498_sample33.csv.gz
28M 9 11 00:22 GSM5573499_sample34.csv.gz
11M 9 11 00:22 GSM5573500_sample35.csv.gz
7.2M 9 11 00:22 GSM5573501_sample36.csv.gz
802K 9 11 00:22 GSM5573502_sample37.csv.gz
5.0M 9 11 00:22 GSM5573503_sample38.csv.gz
10M 9 11 00:22 GSM5573504_sample39.csv.gz
10M 9 11 00:22 GSM5573505_sample40.csv.gz
这个时候千万不要解压它们哦!直接批量读取即可,代码如下所示:
rm(list = ls())
options(stringsAsFactors = F)
library(scRNAstat)
library(Seurat)
library(ggplot2)
library(clustree)
library(cowplot)
library(dplyr)
dir='../GSE183904_RAW'
samples=list.files( dir ,pattern = 'gz')
samples
library(data.table)
sceList = lapply(samples,function(pro){
# pro=samples[1]
print(pro)
ct=fread(file.path( dir ,pro),data.table = F)
ct[1:4,1:4]
rownames(ct)=ct[,1]
ct=ct[,-1]
sce=CreateSeuratObject(counts = ct ,
project = gsub('.csv.gz','',strsplit(pro,'_')[[1]][2]),
min.cells = 5,
min.features = 300,)
return(sce)
})
names(sceList)
library(stringr)
samples=gsub('.csv.gz','',str_split(samples,'_',simplify = T)[,2])
samples
names(sceList) = samples
sceList
我看了看,目前耗费不到10G的内存,小菜一碟啦!
如果这个时候,按照我们的标准单细胞数据处理流程,应该是做harmony或者CCA的整合,然后降维聚类分群,如果你对单细胞数据分析还没有基础认知,可以看基础10讲:
- 01. 上游分析流程
- 02.课题多少个样品,测序数据量如何
- 03. 过滤不合格细胞和基因(数据质控很重要)
- 04. 过滤线粒体核糖体基因
- 05. 去除细胞效应和基因效应
- 06.单细胞转录组数据的降维聚类分群
- 07.单细胞转录组数据处理之细胞亚群注释
- 08.把拿到的亚群进行更细致的分群
- 09.单细胞转录组数据处理之细胞亚群比例比较
但是不符合我们的主题:快速检查迄今为止时间上最大的胃癌单细胞队列数据质量
其实前些天我们在《生信技能树》公众号的一个教程:这也能画?,我提到了一个很无聊的R包,名字是:scRNAstat ,它可以4行代码进行单细胞转录组的降维聚类分群,其实完全没有技术含量, 就是把 Seurat 流程的一些步骤包装成为了4个函数:
- basic_qc (查看数据质量)
- basic_filter (进行一定程度的过滤)
- basic_workflow (降维聚类分群)
- basic_markers(检查各个亚群的标记基因)
这个时候就可以借助它,来快速检查迄今为止时间上最大的胃癌单细胞队列数据质量!
接下来就是见证奇迹的时刻啦, 仅仅是需要不到20行代码,就可以批量完成全部的单细胞样品的各自独立的降维聚类分群的检验!
lapply(names(sceList) , function(x){
# x=names(sceList)[1]
print(x)
sce=sceList[[x]]
sce
dir.create( x )
sce = basic_qc(sce=sce,org='human',
dir = x)
sce
sce = basic_filter(sce)
sce = basic_workflow(sce,dir = x)
markers_figures <- basic_markers(sce,
org='human',
group='seurat_clusters',
dir = x)
p_umap = DimPlot(sce,reduction = 'umap',
group.by = 'seurat_clusters',
label.box = T, label = T,repel = T)
p=p_umap+markers_figures[[1]]
print(p)
ggsave(paste0('umap_markers_for_',x,'.pdf'),width = 12,height = 9)
#save(p,file = paste0('umap_markers_for_',x,'.Rdata'))
})
对每个样品我都快速完成了各自独立的降维聚类分群的检验,而且保存了图片以及图片背后的数据哦!
如果你的内存比较小,也可以把前面的读取步骤跟下面的降维聚类分群步骤合并,这样无需存储每次单细胞数据对象啦!
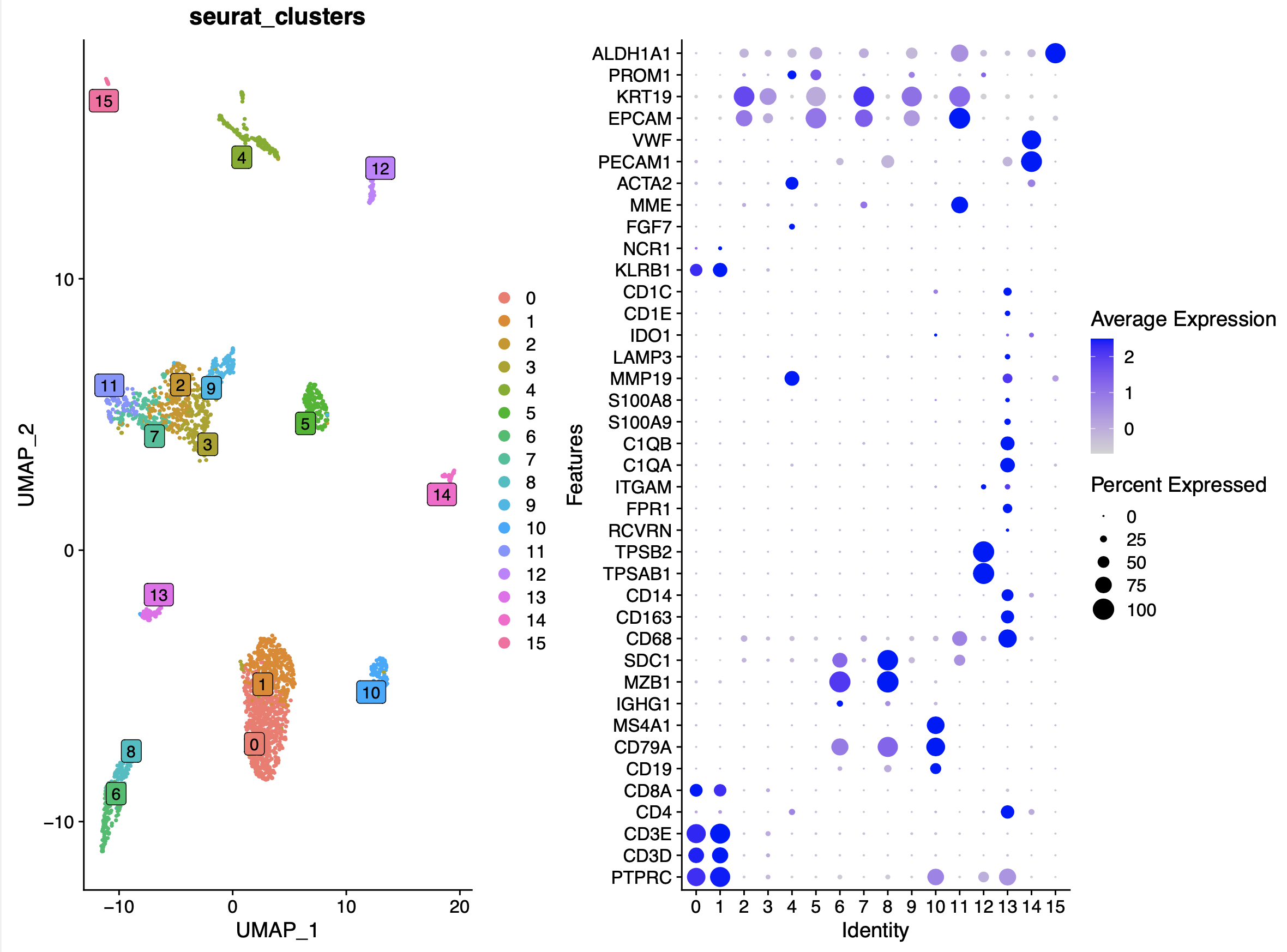
而且只需要背诵如下所示各个细胞亚群高表达量基因的列表:
# T Cells (CD3D, CD3E, CD8A),
# B cells (CD19, CD79A, MS4A1 [CD20]),
# Plasma cells (IGHG1, MZB1, SDC1, CD79A),
# Monocytes and macrophages (CD68, CD163, CD14),
# NK Cells (FGFBP2, FCG3RA, CX3CR1),
# Photoreceptor cells (RCVRN),
# Fibroblasts (FGF7, MME),
# Endothelial cells (PECAM1, VWF).
# epi or tumor (EPCAM, KRT19, PROM1, ALDH1A1, CD24).
# immune (CD45+,PTPRC), epithelial/cancer (EpCAM+,EPCAM),
# stromal (CD10+,MME,fibo or CD31+,PECAM1,endo)
就可以很容易给每个单细胞转录组样品的各个细胞亚群进行生物学命名!
另外,为了感谢大家的付费支持,我把本次数据分析内容打包在腾讯微云啦,只需要在咱们《生信技能树》公众号后台回复「胃癌单细胞」这样的关键词,就可以获取全部的数据和代码,以及运行好的结果哦!还包括文献本身!可以说是超级的人性化啦!
如果你对单细胞数据分析还没有基础认知,可以看基础10讲:
- 01. 上游分析流程
- 02.课题多少个样品,测序数据量如何
- 03. 过滤不合格细胞和基因(数据质控很重要)
- 04. 过滤线粒体核糖体基因
- 05. 去除细胞效应和基因效应
- 06.单细胞转录组数据的降维聚类分群
- 07.单细胞转录组数据处理之细胞亚群注释
- 08.把拿到的亚群进行更细致的分群
当然了,这样的快速检查迄今为止时间上最大的胃癌单细胞队列数据质量并不是真正的单细胞数据分析啦,我们有足够能力对这个数据集进行各式各样的探索,但是苦于没有很好的生物学医学背景,这样的数据宝藏对我们来说简直就是暴殄天物!
如果亲爱的读者,你希望使用这个数据来辅助你的科研想法,请不要犹豫,赶快练习我们吧,仅需800倒1600,我们就会根据你的需求,仅需一定程度的对这个数据集的探索,差异分析或者细胞亚群比例分析,或者更多高级分析。
转录因子分析和细胞通讯分析属于单细胞数据分析里面的高级分析了,主要是对计算机资源消耗会比较大,我也多次分享过细节教程: