看到了预印本出来了一个测评文章;《Comparative Analysis of Commercial Single-Cell RNA Sequencing Technologies》重要的是3个链接:
- Pre-print: https://www.biorxiv.org/content/10.1101/2024.06.18.599579v1
- SRA BioProject: PRJNA1106903 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1106903)
- https://github.com/danledinh/scRNAseq-repo
统一使用了人类的血液样品去做单细胞转录组,涉及到如下所示8种不同的商业化平台:
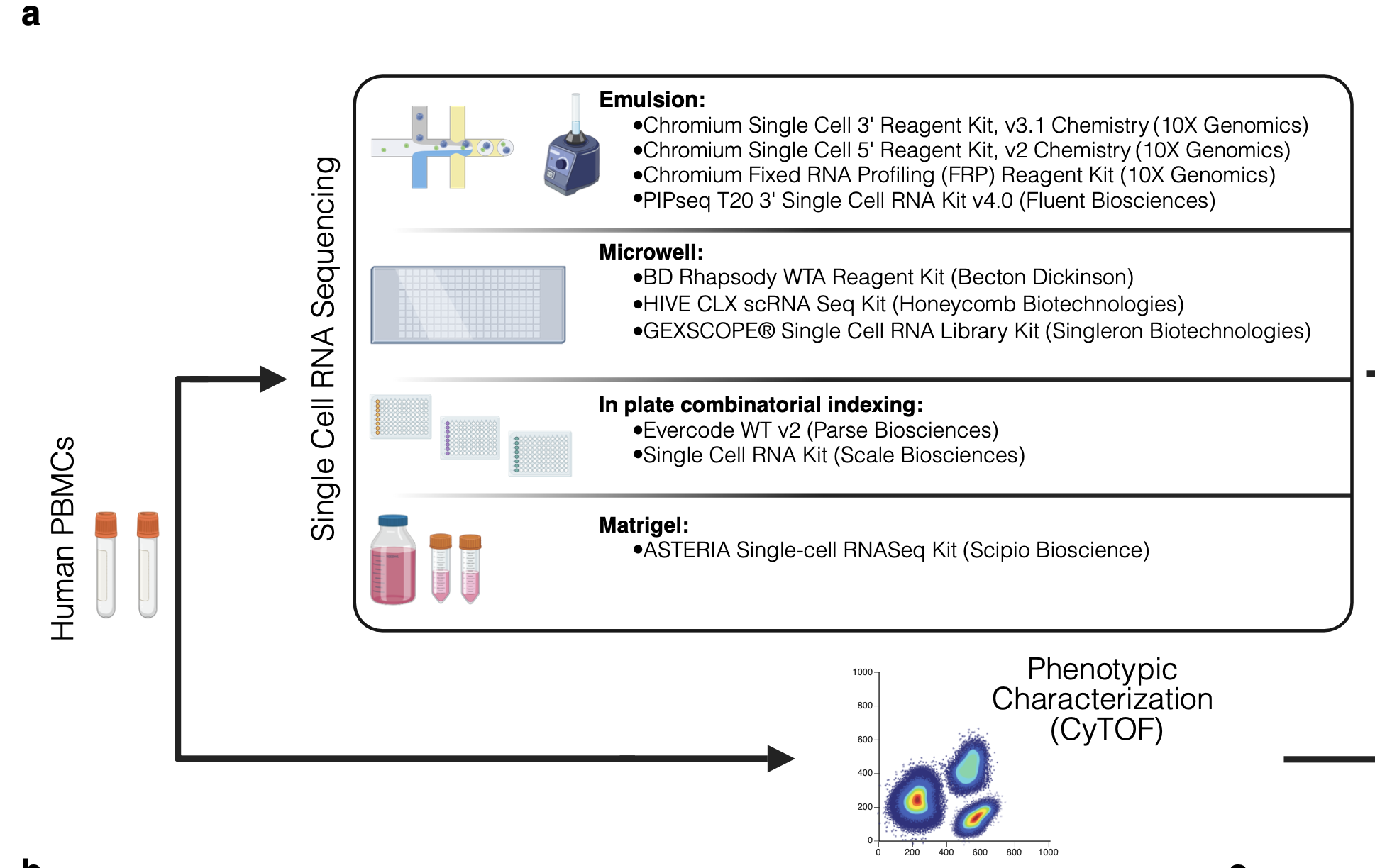
每个商业化单细胞转录组平台都有自己的上游数据处理软件,比如大名鼎鼎的10x就是CellRanger流程,正常走cellranger的定量流程即可,代码我已经是多次分享了。参考:
- 10X单细胞转录组原始测序数据的Cell Ranger流程(仅需800元)
- 10X的单细胞转录组原始数据也可以在EBI下载
- 一个10x单细胞转录组项目从fastq到细胞亚群
- 一文打通单细胞上游:从软件部署到上游分析
- PRJNA713302这个10x单细胞fastq实战
- 一次曲折且昂贵的单细胞公共数据获取与上游处理
- 只能下载bam文件的10x单细胞转录组项目数据处理
- 不知道10x单细胞转录组样品和fastq文件的对应关系
- 10X单细胞转录组测序数据的 SRA转fastq踩坑那些事
- 10x的单细胞转录组fastq文件的R1和R2不能弄混哦
差不多几个小时就可以完成全部的样品的cellranger的定量流程。但是其它平台,因为确实是市场占有率很低,我遇到的不多,所以基本上就只有BD的以及Singleron我分享过实战笔记,但是这个测评文章太强了,https://github.com/danledinh/scRNAseq-repo
- For each of the 10x kits, CellRanger version 7.1.0 was used. The 10x 3’ and 5’ kitsrequired the subcommand count and 10x FRP required the subcommand multi.
- For Fluent,PIPseeker version 2.1 was used.
- For BD, Rhapsody version 2.0 was used.
- For Honeycomb,BeeNet version 1.1.3 was used.
- For Parse, split-pipe version 1.1.1 was used.
- For Scale,ScaleRna version 1.3.3 was used.
- For Singleron, CeleSCOPE version 1.17.0 was used with thesubcommand multi_rna.
- For Scipio, Cytonaut version 7 was used.
The human reference(Ensembl 98/GENCODE v32) from 10x Genomics (refdata-gex-GRCh38-2020-A) was used For alignment and feature quantification.
学徒作业
下载这些样品的fastq文件,然后摸索全部的定量流程:
BD_1 39.31 Gb
BD_2 48.47 Gb
10X_3p_1 7.97 Gb
10X_3p_2 16.67 Gb
10X_5p_1 19.86 Gb
10X_5p_2 11.67 Gb
10X_FRP_1 26.23 Gb
10X_FRP_2 17.11 Gb
Fluent_1 9.67 Gb
Fluent_1 37.63 Gb
Fluent_3 20.10 Gb
Honeycomb_1 9.43 Gb
Honeycomb_2 8.96 Gb
Parse_1 8.97 Gb
Scale_1 14.36 Gb
Scipio_1 8.76 Gb
Scipio_2 5.50 Gb
Singleron_1 73.70 Gb
Singleron_2 77.15 Gb