Monthly Archives: 8月 2015
2014-4742samples-21tumors-Cancer5000-set-254-genes
2015-MADGiC-identify-cancer-driver-gene
2014-REVIEW-identifying driver mutation in sequenced cancer genome
2014-review-Next-generation sequencing to guide cancer therapy
This reductionist thinking led the initial theories on carcinogenesis to be centered on how many “hits” or genetic mutations were necessary for a tumor to develop.
文献笔记-2015-nature-molecular analysis of gastric cancer新的分类及预后调查
文献:Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes
A small pre-defined set of gene expression signatures
| epithelial-to-mesenchymal transition (EMT) | 上皮细胞向间充质细胞转化 |
| microsatellite instability (MSI) | 微卫星不稳定性 |
| cytokine signaling | 细胞因子信号 |
| cell proliferation | 细胞增殖 |
| DNA methylation | DNA甲基化 |
| TP53 activity | TP53活性 |
| gastric tissue | 胃组织 |
经典的分类方法是:Gastric cancer may be subdivided into 3 distinct subtypes—proximal, diffuse, and distal gastric cancer—based on histopathologic and anatomic criteria. Each subtype is associated with unique epidemiology.
我们用主成分分析Principal component anaylsis (PCA)
PC1
PC2
PC3
这三个主成分与上面的七个特征是相关联的。
根据我们的主成分分析,可以把我们的300个GC样本分成如下四组,命名如下:
Gene expression signatures define four molecular subtypes of GC:
MSI (n = 68),
MSS/EMT (n = 46),
MSS/TP53+ (n = 79)
MSS/TP53− (n = 107)
然后用本文的分类方法,测试了另外另个published数据,还是分成四个组
(MSI, MSS/EMT, MSS/TP53+ and MSS/TP53-)
分别是TCGA数据库的;n = 46, n = 62, n = 50 and n = 47.
Singapore的研究; n = 12, n = 85, n = 39 and n = 63 respectively
我们这样的分组可以得到一些规律:
(i) The MSS/EMT subtype occurred at a significantly younger age (P = 3e-2) than did other subtypes. The majority (>80%) of the subjects in this subtype were diagnosed with diffuse-type (P < 1e-4) at stage III/IV(P = 1e-3).
(ii) The MSI subtype occurred predominantly in the antrum (75%), >60% of subjects had the intestinal subtype, and >50% of subjects were diagnosed at an early stage (I/II).
(iii) Epstein-Barr virus (EBV) infection occurred more frequently in the MSS/TP53+ group (n = 12/18, P = 2e-4) than in the other groups.
然后我们对我们的300个样本做了生存分析:
预后: MSI > MSS/TP53+ > MSS/TP53 > MSS/EMT
Next, we validated the survival trend of GC subtypes in three independent cohorts: Samsung Medical Center cohort 2 (SMC-2,n = 277, GSE26253)31,
Singapore cohort(n = 200, GSE15459)21 and
TCGA gastric cohort (n = 205).
We saw that the GC subtypes showed a significant association with overall survival
结论:我们这样的分类是最合理的,跟各个类别的预后非常相关。
然后我们看看突变模式:
the MSI~ hypermutation ~KRAS (23.3%), the PI3K-PTEN-mTOR pathway (42%), ALK (16.3%) and ARID1A (44.2%)18.
We observed enrichment of PIK3CA H1047R mutations in the MSI samples
we saw enrichment of E542K and E545K mutations in MSS tumors
The EMT subtype had a lower number of mutation events when compared to the other MSS groups(P = 1e−3).
The MSS/TP53− subtype showed the highest prevalence of TP53 mutations (60%), with a low frequency of other mutations
the MSS/TP53+ subtype showed a relatively higher prevalence (compared to MSS/TP53−) of mutations in APC, ARID1A, KRAS, PIK3CA and SMAD4.
再看看拷贝数变异情况:
再看看与另外两个研究团队的分类情况的比较
The TCGA study reported expression clusters (subtypes named C1–C4) and genomic subtypes (subtypes named EBV+, MSI, Genome Stable (GS) and Chromosomal Instability (CIN)).
A follow-up study of the Singapore cohort21 described three expression subtypes (Proliferative, Metabolic and Reactive)
However, a consensus on clinically relevant subtypes that encompasses molecular heterogeneity and that can be used in preclinical and clinical research has not been reported.
Here we report the molecular classification of GC linked not only to distinct patterns of genomic alterations, but also to recurrence pattern and prognosis across multiple GC cohorts.
microsatellite instability
英文简称 : MI
中文全称 : 微卫星不稳定性
所属分类 : 生物科学
词条简介 : 微卫星不稳定性(microsatellite instability,MI)检测是基于VNTR的发现,细胞内基因组含有大量的碱基重复序列,一般将6-7bp的串联重称为小卫星DNA(minisatellite DNA),又称为VNTR。而将1-4bp的串联重复称为微卫星DNA,又称简单重复序列(simple repeat sequence,SRS)。SRS是一种最常见的重复序列之一,具有丰富的多态性、高度杂合性、重组纺低等优点。最常见的为双核苷酸重复,即(AC)n和(TG)n。研究表胆,在n≥104时,2bp重复序列在人群中呈高度多态性。SRS广泛存在于原核和真核基因组中,约占真核基因组的5%,是近年来快速发展起来的新的DNA多态性标志之一。策卫星稳定性(MI)是指简重复序列的增加或丢失。MI首先在结肠癌中观察到,1993年在HNPCC中观察到多条染色体均有(AC)n重复序列的增加或毛失,以后相继在胃癌、胰腺癌、肺癌、膀胱癌、乳腺癌、前列腺癌及其他肿瘤等也好现存在微卫星不稳定现象,提示MI可能是肿瘤细胞的另一重要分子结果显示 ,MI与肿瘤与发展有关,MI仅在肿瘤细胞中发现,从未在正常组织中检测到。在原发与移肿瘤中,MI均交分布于整个肿瘤。晚期胃癌的MI频率显著高于早期胃癌。
文献笔记-2010-R-softeware-identify-cancer_driver_genes
我们用188 non-small cell lung tumors数据来测试了一个R语言程序,find driver genes in cancer ~
软件地址如下:http://linus.nci.nih.gov/Data/YounA/software.zip
这是一个R语言程序,里面有readme,用法很简单。
准备好两个文件,分别是silent_mutation_table.txt and nonsilent_mutation_table.txt ,它们都是普通文本格式数据,内容如下,就是把找到的snp格式化,根据注释结果分成silent和nonsilent即可。
#Ensembl_gene_id Chromosome Start_position Variant_Type Reference_Allele Tumor_Seq_Allele1 Tumor_Seq_Allele2 Tumor_Sample_Barcode
#ENSG00000122477 1 100390656 SNP G G A TCGA-23-1022-01A-02W-0488-09
然后直接运行程序包里面的主程序,在R语言里面source("main_R_script.r")
We reanalyzed sequence data for 623 candidate genes in 188 non-small cell lung tumors using the new method.
to identify genes that are frequently mutated and thereby are expected to have primary roles in thedevelopment of tumor
To find these driver genes, each gene is tested for whether its mutation rate is significantly higher than the background (or passenger) mutation rate.
Some investigators (Sjoblom et al., 2006) further divide mutations into several types according to the nucleotide and the neighboring nucleotides of the mutations.
Ding et al. (2008)的方法的三个缺点:
1、different types of mutations can have different impact on proteins.(越影响蛋白功能的突变,越有可能是driver mutation)
2、different samples have different background mutation rates. (在高突变背景的样本中的突变,很可能是高突变背景的原因,而不是因为癌症)
3、a different number of non-silent mutations can occur at each base pair according to the genetic code.(比如Tryptophan仅仅只有一个密码子,而arginine高达6个密码子)
我们提出的方法的4个优点是:
1,我们对非同义突变根据它们对蛋白功能的影响进行了评级打分。
2,我们允许不同的样品有着不同的BMR
3,that whether the mutation is non-silent or silent depends on the genetic code
4,we take into account uncertainties in the background mutation rate by using empirical Bayes methods
还有5个需要改进的地方:
1,However, the functional impact is also dependent on the position in which a mutation occurs.(我们仅仅考虑了突变对氨基酸的改变)
2,the current scoring system which assigns mutation scores in the order: missense mutation<inframe indel<mutation in splice sites<frameshift indel=nonsense mutation may be biased toward identifying tumor suppressor genes over oncogenes.
3,we may refine our background mutation model in Table 1 so that all six types of mutations, A:T→G:C, A:T→C:G, A: T→T :A,G:C→A:T, G:C→T :A, G:C→C:G have separate mutation rates.
4,we did not take into account correlations among mutations in identifying driver genes.
5,one might combine both copy number variation and sequencing data to identify driver genes.
HGNC定义的gene Symbol转为ensemble数据库的ID,的R语言代码:
library(biomaRt)
ensembl=useMart("ensembl",dataset = "hsapiens_gene_ensembl")
all.gene.table = read.table("all_gene.symbol", header=F)
convert=getBM(attributes = c("chromosome_name","ensembl_gene_id","hgnc_symbol"),filters =c("hgnc_symbol"),values=all.gene.table[,1],mart=ensembl)
chromosome=c(1:22,"X","Y","M")
convert=convert[!is.na(match(convert[,1],chromosome)),2:3] #remove names whose matching chromosome is not 1-22, X, or Y.
convert=convert[rowSums(convert=="")==0,]
write.table(convert,"ensembl2symbol.list",quote = F,row.names =F,col.names =F)
write.table(convert,"all_gene_name.txt",quote = F,row.names =F,col.names =F)
一个gene Symbol可能对应着多个ensemble ID号,但是在每个染色体上面是一对一的关系。
有些gene Symbol可能找不到ensemble ID号,一般情况是因为这个gene Symbol并不是纯粹的HGNC定义的,或者是比较陈旧的ID。
比如下面的TIGAR ,就很可能被写作是C12orf5
Aliases for TIGAR Gene
TP53 Induced Glycolysis Regulatory Phosphatase 2 3
TP53-Induced Glycolysis And Apoptosis Regulator 2 3 4
C12orf5 3 4 6
Probable Fructose-2,6-Bisphosphatase TIGAR 3
Fructose-2,6-Bisphosphate 2-Phosphatase 3
Chromosome 12 Open Reading Frame 5 2
Fructose-2,6-Bisphosphatase TIGAR 3
Transactivated By NS3TP2 Protein 3
EC 3.1.3.46 4
FR2BP 3
External Ids for TIGAR Gene
HGNC: 1185 Entrez Gene: 57103 Ensembl: ENSG00000078237 OMIM: 610775 UniProtKB: Q9NQ88
Previous HGNC Symbols for TIGAR Gene
C12orf5
Export aliases for TIGAR gene to outside databases
研究癌症领域必看文献
最近需要了解一些癌症相关知识,看到了这个文献列表,觉得非常棒,所以推荐给大家。
抽时间慢慢看,一个月应该可以把这些文献看完的。
癌症种类大全 http://www.cancer.gov/types
癌症药物大全 http://www.cancer.gov/about-cancer/treatment/drugs
癌症所有的信息几乎都能在这个网站上面找到 http://www.cancer.gov/
包括癌症的科普、treatment、diagnosis,prognosis,classification,drugs、prediction等等
Cancer Precision Medicine: Improving Evidence in Practice - August 24, 2015
NCI-MATCH Trial Opens,![]() AACR blog post, August 2015
AACR blog post, August 2015
NCI-MATCH launch highlights new trial design in precision-medicine era![]()
McNeal C , JNCI, August 2015
The Cancer Genomics Resource List, 2014![]()
Zutter MM et al. CAP Lab Improvement Program,Archives of Pathology, August 2015
Personalized medicine and economic evaluation in oncology: all theory and no practice?![]()
Garattini L et al. Expert Rev Pharmacoecon Outcomes Res 2015 Aug 9. 1-6
Precision medicine trials bring targeted treatments to more patients,![]() C. Helwick, ASCO Post, Jul 25
C. Helwick, ASCO Post, Jul 25
Next-generation sequencing to guide cancer therapy ![]()
Gagan J et al, Genome Medicine, July 29, 2015
Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials.![]()
Meric-Bernstam F et al. J. Clin. Oncol. 2015 May 26.
Brave-ish new world-what's needed to make precision oncology a practical reality.![]()
MacConaill LE et al. JAMA Oncol 2015 Jul 16.
Genomic profiling: Building a continuum from knowledge to care![]()
Helen C et al. JAMA Oncology, July 2015
Are we there yet?![]()
When it comes to curing cancer, targeted therapies and genomic sequencing are helping, but we still have far to go. Genome Magazine, June 29, 2015
Artificial intelligence, big data, and cancer![]()
Kantarjian H et al, JAMA Oncology, June 2015
Multigene panel testing in oncology practice - how should we respond?![]()
Kurian AW et al. JAMA Oncology, June 2015
Use of whole genome sequencing for diagnosis and discovery in the cancer genetics clinic.![]()
Foley SB et al. EBioMedicine 2015 Jan 2(1) 74-81
The future of molecular medicine: biomarkers, BATTLEs, and big data ![]()
ES Kim, ASCO University, June 2015
NCI-MATCH trial will link targeted cancer drugs to gene abnormalities![]()
Targeted agent and profiling utilization registry study,![]() from the American Society for Clinical Oncology
from the American Society for Clinical Oncology
ASCO study aims to learn from patient access to targeted cancer drugs used off-label,![]() American Society for Clinical Oncology
American Society for Clinical Oncology
Improving evidence developed from population-level experience with targeted agents ![]() [PDF 462.93 KB]
[PDF 462.93 KB]![]()
McLellan M et al Issue Brief. Conference on Clinical Cancer Research November 2014
Implementing personalized cancer care.![]()
Schilsky RL et al. Nat Rev Clin Oncol 2014 Jul (7) 432-8
Accelerating the delivery of patient-centered, high-quality cancer care.![]()
Abrahams E et al. Clin. Cancer Res. 2015 May 15. (10) 2263-7
Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity.![]()
Catenacci DV et al. Mol Oncol 2015 May (5) 967-996
Cancer Precision Medicine: Improving Evidence in Practice - May 29, 2015
Diagnosis and treatment of cancer using genomics![]()
Vockley JG et al. BMJ, May 28, 2015
Targeted agent and profiling utilization registry study,![]() from the American Society for Clinical Oncology
from the American Society for Clinical Oncology
ASCO study aims to learn from patient access to targeted cancer drugs used off-label,![]() American Society for Clinical Oncology
American Society for Clinical Oncology
Improving evidence developed from population-level experience with targeted agents ![]() [PDF 462.93 KB]
[PDF 462.93 KB]![]()
McLellan M et al Issue Brief. Conference on Clinical Cancer Research November 2014
Implementing personalized cancer care.![]()
Schilsky RL et al. Nat Rev Clin Oncol 2014 Jul (7) 432-8
Accelerating the delivery of patient-centered, high-quality cancer care.![]()
Abrahams E et al. Clin. Cancer Res. 2015 May 15. (10) 2263-7
Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity.![]()
Catenacci DV et al. Mol Oncol 2015 May (5) 967-996
Precision Medicine: Cancer and Genomics - May 12, 2015
Promise, peril seen in personalized cancer therapy,![]() by Marie McCullough, Philadelphia Inquirer, May 10
by Marie McCullough, Philadelphia Inquirer, May 10
A decision support framework for genomically informed investigational cancer therapy.![]()
Meric-Bernstam F et al. J. Natl. Cancer Inst. 2015 Jul (7)
Divide and conquer: The molecular diagnosis of cancer,![]() by Louis M. Staudt, National Cancer Insitute, Apr 13
by Louis M. Staudt, National Cancer Insitute, Apr 13
Health: Make precision medicine work for cancer care![]()
To get targeted treatments to more cancer patients pair genomic data with clinical data, and make the information widely accessible, Mark A. Rubin. Nature News, Apr 15
Using somatic mutations to guide treatment decisions![]()
Horlings H et al. JAMA Oncology, March 12, 2015
The landscape of precision cancer medicine clinical trials in the United States![]()
Roper N et al. Cancer Treatment Reviews 2015
What is “precision medicine?![]() Information from the National Cancer Institute
Information from the National Cancer Institute
Impact of cancer genomics on precision medicine for the treatment of cancer,![]() from the Cancer Genome Atlas, NCI
from the Cancer Genome Atlas, NCI
US precision-medicine proposal sparks questions,![]() by Sara Reardon, Nature News, Jan 22
by Sara Reardon, Nature News, Jan 22
Obama's 'precision medicine' means gene mapping,![]() NBC News, Jan 21
NBC News, Jan 21
What is President Obama's 'precision medicine' plan, and how might it help you?![]() By Lenny Bernstein, Jan 21
By Lenny Bernstein, Jan 21
Recent reviews
Companion diagnostics: the key to personalized medicine.![]()
Jørgensen JT. Expert Rev Mol Diagn. 2015 Feb;15(2):153-6
Promoting precision cancer medicine through a community-driven knowledgebase.![]()
Geifman N, et al. J Pers Med. 2014 Dec 15;4(4):475-88.
Toward a prostate cancer precision medicine.![]()
Rubin MA. Urol Oncol. 2014 Nov 20.
Prioritizing targets for precision cancer medicine.![]()
Andre F, et al. Ann Oncol. 2014 Dec;25(12):2295-303
Toward precision medicine with next-generation EGFR inhibitors in non-small-cell lung cancer.![]()
Yap TA, Popat S. Pharmgenomics Pers Med. 2014 Sep 19;7:285-95.
Genomically driven precision medicine to improve outcomes in anaplastic thyroid cancer.![]()
Pinto N, et al. J Oncol. 2014;936285
Translating genomics for precision cancer medicine.![]()
Roychowdhury S, Chinnaiyan AM. Annu Rev Genomics Hum Genet. 2014;15:395-415
The Cancer Genome Atlas: Accomplishments and Future - April 3, 2015
The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge![]()
Tomczak K, et al. Contemp Oncol (Pozn). 2015; 19(1A): A68-A77.
The Cancer Genome Atlas' 4th Annual Scientific Symposium![]()
May 11-12 ~ Bethesda, MD
The Cancer Genome Atlas (TCGA) Data Portal ![]()
Portal provides a platform for researchers to search, download, and analyze data sets generated by TCGA
Cancer Genomics Hub: A resource of the National Cancer Institute,![]() from the USC Genome Browser
from the USC Genome Browser
Molecular classification of gastric adenocarcinoma: translating new insights from The Cancer Genome Atlas Research Network.![]()
Sunakawa Y et al. Curr Treat Options Oncol 2015 Apr (4) 331
TCGA data and patient-derived orthotopic xenografts highlight pancreatic cancer-associated angiogenesis.![]()
Gore J et al. Oncotarget 2015 Feb 25.
Radiogenomics of clear cell renal cell carcinoma: preliminary findings of The Cancer Genome Atlas-Renal Cell Carcinoma (TCGA-RCC) Imaging Research Group.![]()
Shinagare AB et al. Abdom Imaging 2015 Mar 10.
Proteomics of colorectal cancer in a genomic context: First large-scale mass spectrometry-based analysis from the Cancer Genome Atlas.![]()
Jimenez CR et al. Clin. Chem. 2015 Feb 26.
End of cancer-genome project prompts rethink![]()
Geneticists debate whether focus should shift from sequencing genomes to analysing function. Heidi Ledford, Nature News and Comments, January 2015
Cancer Genomics: Insights into Driver Mutations - March 10, 2015
Seek and destroy: Relating cancer drivers to therapies![]()
E. Martinez-Ledesma et al. Cell, March 9, 2015
In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities![]()
C Rubio-Perez et al. Cancer Cell, March 9, 2015
MADGiC: a model-based approach for identifying driver genes in cancer. ![]() [PDF 373.56 KB]
[PDF 373.56 KB]![]()
Keegan D. Korthauer et al. Bioinformatics, January 2015
Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine.![]()
Benjamin J Raphael et al. Genome Medicine 2014
Novel recurrently mutated genes in African American colon cancers.![]()
Guda K et al. Proc Natl Acad Sci U S A. 2015 Jan 12
Sparse expression bases in cancer reveal tumor drivers.![]()
Logsdon BA, et al. Nucleic Acids Res. 2015 Jan 12
Patient-specific driver gene prediction and risk assessment through integrated network analysis of cancer omics profiles.![]()
Bertrand D, et al. Nucleic Acids Res. 2015 Jan 8
Identification of constrained cancer driver genes based on mutation timing.![]()
Sakoparnig T, et al. PLoS Comput Biol. 2015 Jan 8;11(1):e1004027
CaMoDi: a new method for cancer module discovery.![]()
Manolakos A, et al. BMC Genomics. 2014 Dec 12;15 Suppl 10:S8.
VHL, the story of a tumour suppressor gene.![]()
Gossage L, et al. Nat Rev Cancer. 2014 Dec 23;15(1):55-64
Targeting the MET pathway for potential treatment of NSCLC.![]()
Li A, et al. Expert Opin Ther Targets. 2014 Dec 23:1-12
Deciphering oncogenic drivers: from single genes to integrated pathways.![]()
Chen J, et al. Brief Bioinform. 2014 Nov 5.
Driver and passenger mutations in cancer.![]()
Pon JR, et al. Annu Rev Pathol. 2014 Oct 17
Hereditary Cancer Genetic Testing: Where are We? - December 18, 2014
NCI paper:Prevalence and correlates of receiving and sharing high-penetrance cancer genetic test results: Findings from the Health Information National Trends Survey![]()
Taber J.M. et al Public Health Genomics, January 2015
Clinical decisions: Screening an asymptomatic person for genetic risk--polling results![]()
Schulte J, et al. N Engl J Med 2014 Nov;371(20):e30
Testing for hereditary breast cancer: Panel or targeted testing? Experience from a clinical cancer genetics practice.![]()
Doherty J, J Genet Couns. 2014 Dec 5
Hereditary colorectal cancer syndromes: American Society of Clinical Oncology clinical practice guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology clinical practice guidelines.![]()
Stoffel EM, et al. J Clin Oncol. 2014 Dec 1
Population testing for cancer predisposing BRCA1/BRCA2 mutations in the Ashkenazi-Jewish community: A randomized controlled trial.![]()
Manchanda R, et al. J Natl Cancer Inst. 2014 Nov 30;107(1)
Cost-effectiveness of population screening for BRCA mutations in Ashkenazi Jewish women compared with family history-based testing.![]()
Manchanda R et al. J Natl Cancer Inst. 2014 Nov 30;107(1). pii: dju380. doi: 10.1093/jnci/dju380. Print 2015 Jan.
Check out our Cancer Genetic Testing Update Page for additional information and links
Cancer Genomic Tests (October 30, 2014)
Cancer Genomic Tests: Accelerating Translation - October 30, 2014
CDC-NCI paper: An overview of recommendations and translational milestones for genomic tests in cancer![]()
Christine Q. Chang et al. Genetics in Medicine, October 22, 2014
Check out the CDC evidence-based classification of cancer genomic tests
Check out the NCI Cancer Genomics and Epidemiology Navigator for latest information on cancer genomic tests![]()
EGAPP: A model process for evaluating genomic applications in practice and prevention.![]() Check out cancer genomic tests, methods, evidence reviews and recommendation statements.
Check out cancer genomic tests, methods, evidence reviews and recommendation statements.
NCI Fact Sheet: Genetic testing for hereditary cancer syndromes![]()
Cancer Genomics: Impact of Recent Insights - October 30, 2014
Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin.![]()
Katherine A. Hoadley et al. Cell, August 2014
Genome study overhauls cancer categories, shifts from tissues to molecular subtypes,![]() by Kevin Mayer, Genetics Bioengineering News, Aug 8
by Kevin Mayer, Genetics Bioengineering News, Aug 8
It's time for us to think about cancer differently,![]() by Paula Mejia, Newsweek, Aug 8
by Paula Mejia, Newsweek, Aug 8
NIH- The Cancer Genome Atlas (TCGA) Initiative![]()
NIH information: What is cancer genomics and the genetic basis of cancer?![]()
Cancer Precision Medicine: Where Are We? - September 18, 2014
NIH announces the launch of 3 integrated precision medicine trials; ALCHEMIST is for patients with certain types of early-stage lung cancer,![]() August 2014
August 2014
National Cancer Institute's Precision Medicine Initiatives for the New National Clinical Trials Network.![]() Jeffrey Abrams et al. ASCO Annual Meeting 2014
Jeffrey Abrams et al. ASCO Annual Meeting 2014
Personalized medicine: Special treatment.![]()
Michael Eisenstein. Nature, September 11, 2014
Why the controversy? Start sequencing tumor genes at diagnosis. Tumor sequencing at the time of diagnosis can give significant insight for successful cancer treatment,![]() by Shelly Gunn, Genetic Engineering & Biotechnology News, Sep 10
by Shelly Gunn, Genetic Engineering & Biotechnology News, Sep 10
National Cancer Institute information: Precision medicine and targeted therapy![]()
Genomics and precision oncology: What's a targeted therapy for cancer?![]() An updated list of approved drugs from the National Cancer Institute (2014)
An updated list of approved drugs from the National Cancer Institute (2014)
Therapy: This time it's personal![]()
Gravitz L Nature 509, S52-S54 2014 May 29
Multi-marker solid tumor panels using next-generation sequencing to direct molecularly targeted therapies![]()
Michael Marrone, et al. PLoS Currents Evidence on Genomic Tests 2014 May 27
Impact of cancer genomics on precision medicine for the treatment of cancer,![]() from the National Cancer Institute
from the National Cancer Institute
Cancer genomics and precision medicine in the 21st century ![]() [PDF 2.20 MB]
[PDF 2.20 MB]![]() , power point presentation from the National Human Genome Research Institute
, power point presentation from the National Human Genome Research Institute
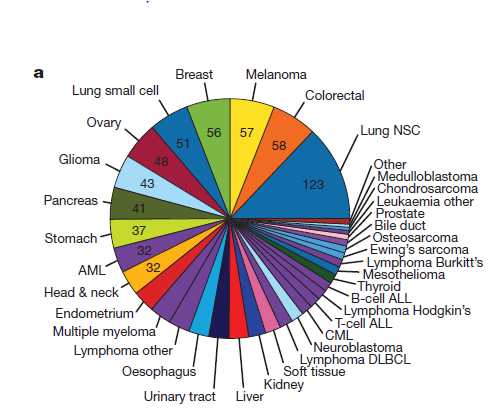
TCGA数据库的癌症种类以及癌症相关基因列表
TCGA projects 里面包含的癌症种类非常多,但是我们分析数据时候常常用pan-cancer 12,pan-cancer 17,pan-cancer 21来表示数据集有多少种癌症,一般文献会给出癌症的简称或者全名:
BLCA, BRCA, COADREAD, GBM, HNSC, KIRC, LAML, LGG, LUAD, LUSC, OV, PRAD, SKCM, STAD, THCA, UCEC.
Acute myeloid leukaemia
Bladder
Breast
Carcinoid
Chronic lymphocytic leukaemia
Colorectal
Diffuse large B-cell lymphoma
Endometrial
Oesophageal adenocarcinoma
Glioblastoma multiforme
Head and neck
Kidney clear cell
Lung adenocarcinoma
Lung squamous cell carcinoma
Medulloblastoma
Melanoma
Multiple myeloma
Neuroblastoma
Ovarian
Prostate
Rhabdoid tumour
HCD features: download
这是高置信度的癌症驱动基因列表:共280多个基因
Cancer5000 features: download
这是一篇对接近5000个癌症样本的研究得到的癌症相关基因列表:共230多个基因
参考:http://bg.upf.edu/oncodrive-role/
http://bioinformatics.oxfordjournals.org/content/30/17/i549.full
http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html?WT.ec_id=NATURE-20140123
TCGA年度研讨会资料分享
TCGA想必搞生信都或有耳闻,尤其是癌症研究方向的,共4个年度研讨会,主要是pdf格式的ppt分享,有需要的可以具体点击到页面一个个下载自己慢慢研究,也可以用我下面链接直接下载。
本来是有youtube分享演讲视频的,但是国内被墙了,大家就看看ppt吧
http://www.genome.gov/17516564
The Cancer Genome Atlas (TCGA) is a comprehensive and coordinated effort to accelerate our understanding of the molecular basis of cancer through the application of genome analysis technologies, including large-scale genome sequencing.
TCGA is a joint effort of the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), which are both part of the National Institutes of Health, U.S. Department of Health and Human Services.
Meetings
- The Cancer Genome Atlas Fourth Annual Scientific Symposium

May 11-12, 2015 - The Cancer Genome Atlas Third Annual Scientific Symposium
May 12-13, 2014 - The Cancer Genome Atlas Second Annual Scientific Symposium
November 27-28, 2012 - The Cancer Genome Atlas First Annual Scientific Symposium
November 17-18, 2011
pdf链接地址如下
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Laird.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Durbin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Ley.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Sartor.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Ciriello.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Imielinski.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Gao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Carter.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Ng.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Parvin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Raphael.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Lawrence.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Kreisberg.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Marra.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Helman.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Stuart.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Cooper.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Levine.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Natsoulis.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Haussler.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Erkkila.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Gehlenborg.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Qiao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Sivachenko.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Sumazin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Gutman.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Mardis.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/01_Shaw.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/02_Chanock.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/03_Staudt.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/05_Creighton.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/06_Stojanov.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/07_Karchin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/08_Mungall.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/09_Hakimi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/10_Gao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/11_Hayes.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/12_Troester.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/13_Knobluach.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/14_Raphael.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/15_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/16_Giordano.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/17_Weinstein.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/18_Zheng.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/19_Getz.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/20_VanDneBroek.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/21_Liao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/22_Khazanov.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/23_Levine.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/24_Miller.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/25_Ewing.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/26_Cirello.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/27_Verhaak.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/28_Hofree.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/29_Meyerson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/30_Yang.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/31_Wheeler.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/32_Parfenov.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/33_Bernard-Rovira.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/34_Hast.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/36_Sellars.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/04_Brat.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/05_Mungall.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/06_Boutros.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/07_Zmuda.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/08_Benz.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/09_Zheng.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/11_Creighton.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/12_Aksoy.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/13_Dinh.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/14_Stuart.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/15_Amin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/16_Gross.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/15_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/18_Giordano.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/19_Amin-Mansour.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/20_Oesper.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/21_Gatza.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/22_Bernard.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/23_Sinha.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/24_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/25_Watson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/26_Martignetti.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/27_Bandlamudi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/28_Fu.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/29_Akdemir.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/30_Bass.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/31_Hakimi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/32_Wheeler.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/33_Lehmann.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/34_Gordenin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/35_Wyczalkowski.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/02_Zenklusen.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/03_Hutter.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/04_Brat.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/05_Mungall.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/06_Linehan.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/07_Brooks.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/08_Wu.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/09_Giger.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/10_Wilkerson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/11_Orsulic.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/12_Zhong.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/13_Knijnenburg.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/14_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/15_Wang.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/16_Poisson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/17_Alaeimahabadi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/18_Noushmehr.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/19_Pantazi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/20_Shih.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/21_Stransky.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/22_Giordano.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/23_Davidsen.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/24_Gross.pdf
R语言实现并行计算
前面我提到有一个大的运算任务需要很久才完成,所以用到了进度条来监控过程,但并不是改善了计算速度,所以需要用到并行计算,我又在网上找了找。
同样也是一个包,跟matlab的实现过程很像
library(parallel)
cl.cores <- detectCores() #检查当前电脑可用核数。
cl <- makeCluster(cl.cores) #使用刚才检测的核并行运算
#这里用clusterEvalQ或者par开头的apply函数族就可以进行并行计算啦
stopCluster(cl)
R-Doc里这样描述makeCluster函数:Creates a set of copies of R running in parallel and communicating over sockets. 即同时创建数个R进行并行运算。在该函数执行后就已经开始并行运算了,电脑可能会变卡一点。尤其在执行par开头的函数时。
在并行运算环境下,常用的一些计算方法如下:
1、clusterEvalQ(cl,expr)函数利用创建的cl执行expr。这里利用刚才创建的cl核并行运算expr。expr是执行命令的语句,不过如果命令太长的话,一般写到文件里比较好。比如把想执行的命令放在Rcode.r里:clusterEvalQ(cl,source(file="Rcode.r"))
2、par开头的apply函数族。这族函数和apply的用法基本一样,不过要多加一个参数cl。一般如果cl创建如上面cl <- makeCluster(cl.cores)的话,这个参数可以直接用作parApply(cl=cl,…)。当然Apply也可以是Sapply,Lapply等等。注意par后面的第一个字母是要大写的,而一般的apply函数族第一个字母不大写。
另外要注意,即使构建了并行运算的核,不使用parApply()函数,而使用apply()函数的话,则仍然没有实现并行运算。换句话说,makeCluster只是创建了待用的核,而不是并行运算的环境。
参考:http://www.r-bloggers.com/lang/chinese/1131
然后我模仿着用并行计算实现自己的需求
#it did work very fast
library(parallel)
cl.cores <- detectCores()
cl <- makeCluster(cl.cores)
clusterExport(cl, "all_dat_t") #这里是重点,因为并行计算里面用到了自定义函数
clusterExport(cl, "all_prob_id") #但是这个函数需要用到这两个数据,所以需要把这两个数据加载到并行计算环境里面
prob_202723_s_at=parSapply( #我这里用的parSapply来实现并行计算
cl=cl, #其中cl是我前面探测到的core数量,
deviation_prob, #deviation_prob是我待并行处理的向量
test_pro #这里其实应该是一个自定义函数,我这里就不写出来了,对上面的deviation_prob向量的每个探针都进行判断
)
R语言实现进度条
我也是临时在网上搜索到的教程,然后简单看了一下就实现了,其实就是就用到了一个名称为tcltk的包,直接查看函数tkProgressBar就可以知道怎么用啦!
下面是网上的一个小的示例代码(么有实际意义,仅为举例而已):
library(tcltk2)
u <- 1:2000
plot.new()
pb <- tkProgressBar("进度","已完成 %", 0, 100)
for(i in u) {
x<-rnorm(u)
points(x,x^2,col=i)
info <- sprintf("已完成 %d%%", round(i*100/length(u)))
setTkProgressBar(pb, i*100/length(u), sprintf("进度 (%s)", info), info)
}
close(pb)#关闭进度条
但是下面的代码是我模仿上面这个教程自己实现的。
[R]
# 以下是实现进度条
library(tcltk2)
plot.new()
pb <- tkProgressBar("进度","已完成 %", 0, 100)
prob_202723_s_at_value=rep(0,length(deviation_prob))
start_time=Sys.time() #这里可以计时,因为要实现进度条的一般都是需要很长运算时间
for (i in 1:length(deviation_prob)) {
tmp=test_pro(deviation_prob[i]) #test_pro是我自定义的一个函数,判断该探针是否符合要求。
if (length(tmp)!=0){prob_202723_s_at_value[i]=tmp}
info <- sprintf("已完成 %d%%", round(i*100/length(deviation_prob))) #进度条就是根据循环里面的i来看看循环到哪一步了
setTkProgressBar(pb, i*100/length(deviation_prob), sprintf("进度 (%s)", info), info)
}
close(pb)#关闭进度条
end_time=Sys.time()
cat(end_time-start_time)
[/R]
R语言-比较数据框提取列的速度
结论:从数据框里面取某列数据,三种方法的时间消耗区别很大,直接用索引值,是最快的,而用$符号其次,用列名最慢。
我在R里面建立了一个表达量矩阵,列名是一个个样品,行是一个个探针,矩阵值是该探针在该样品测定的表达量。
那么,如果我要看看名为"202723_s_at"的探针的表达向量与其它所有探针的表达向量的相关系数,我可以用以下三种方法:
> system.time(apply(all_dat_t,2,function(x) cor(all_dat_t$"202723_s_at",x)))
user system elapsed
22.93 0.03 23.03
> system.time(apply(all_dat_t,2,function(x) cor(all_dat_t[,"202723_s_at"],x)))
Timing stopped at: 92.02 5.32 97.66
太耗时间了,省去
> system.time(apply(all_dat_t,2,function(x) cor(all_dat_t[,grep(prob,names(all_dat_t))],x)))
Timing stopped at: 13.55 0.04 13.66
> prob_num=grep(prob,names(all_dat_t))
> system.time(apply(all_dat_t,2,function(x) cor(all_dat_t[,prob_num],x)))
user system elapsed
8.14 0.01 8.17
可以看出,如果我首先根据探针名,grep出它在该表达量矩阵的列数,然后用列数来提取它的表达量是最快的,而且时间改善非常明显!
我们再探究一下cor函数的效率问题
探究的矩阵有54675个变量,每个变量均有189个观测值,如果取这个大矩阵的部分变量来求相关系数,结果如下!
> system.time(cor(all_dat_t[,1:10]))
user system elapsed
0.001 0.000 0.001
> system.time(cor(all_dat_t[,1:100]))
user system elapsed
0.003 0.000 0.003
> system.time(cor(all_dat_t[,1:1000]))
user system elapsed
0.107 0.002 0.108
> system.time(cor(all_dat_t[,1:10000]))
user system elapsed
11.102 0.849 11.983
> system.time(cor(all_dat_t)) 约六分钟也是可以搞定的
但是如果cor(all_dat_t),六分钟后得到的相关系数矩阵约32G,非常恐怖!
但是它很明显没有把这个32G相关系数矩阵存储到内存,因为我的机器本来就16G内存。我至今不能明白R具体实现机理
生信教程推荐-MSU的一个生信课程
http://angus.readthedocs.org/en/2014/index.html
Next-Gen Sequence Analysis Workshop (2014)
This is the schedule for the 2014 MSU NGS course.
This workshop has a Workshop Code of Conduct.
Download all of these materials or visit the GitHub repository.
| Day | Schedule |
| Monday 8/4 |
|
| Tuesday 8/5 |
|
| Wed 8/6 |
|
| Thursday 8/7 |
|
| Friday 8/8 |
|
| Saturday 8/9 |
|
| Monday 8/11 |
|
| Tuesday 8/12 |
|
| Wed 8/13 |
|
| Thursday 8/14 |
|
| Friday 8/15 |
|
根据基因表达量对样品进行分类ConsensusClusterPlus
bioconductor系列的包都是一样的安装方式:
source("http://bioconductor.org/biocLite.R")
biocLite("ConsensusClusterPlus")
这个包是我见过最简单的包, 加载只有做好输入数据,只需要一句话即可运行,然后默认输出所有结果
> d[1:5,1:5]
d = sweep(d,1, apply(d,1,median,na.rm=T))
从GEO数据库下载矩阵数据-可以直接进行下游分析
bioconductor系列的包都是一样的安装方式:
source("http://bioconductor.org/biocLite.R")
biocLite("GEOquery")
以前GEO数据库主要是microarray的芯片数据,后来有了RNA-seq,如果同时做多个样品的RNA-seq,表达量矩阵后来也可以上传到GEO数据库里面,只有看到文献里面有提到GEO数据库,都可以通过这个R包俩进行批量下载,其实就是网页版的一个API调用而已:
这个函数有很多参数,除非你需要下载的文件,那么就设置destdir到你喜欢的目录,如果只需要表达量数据就不用了。
g4102 <- GDS2eSet(getGEO("GDS4102"))
e4102<-exprs(g4102)
e4100<-exprs(g4100)
#Download GDS file, put it in the current directory, and load it:
gds858 <- getGEO('GDS858', destdir=".")
如果使用了GSEMatrix=TRUE这个参数,那么除了下载soft文件,还有表达量矩阵文件,可以直接用read.table读取那个文件。
#Or, open an existing GDS file (even if its compressed):
gds858 <- getGEO(filename='GDS858.soft.gz')
下面这个下载的是GSE对象,GDS对象还有大一点hpv病毒研究调研
最新文献 http://www.ncbi.nlm.nih.gov/pubmed/26086163 上面有提到了hpv的研究现状
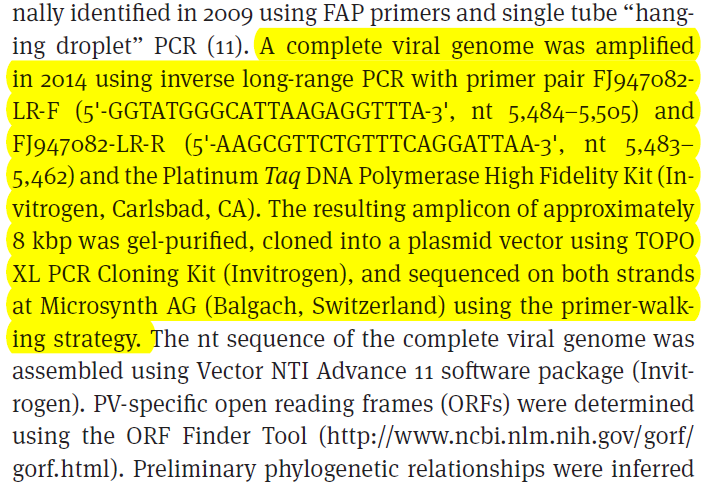
As of May 30, 2015, 201 different HPV types had been completely sequenced and officially recognized and divided into five PV-genera: Alpha-, Beta-, Gamma-, Mu-, and Nupapillomavirus.
根据文献,我找到了hpv所有已知测序种类的参考基因组网站:
http://www.hpvcenter.se/html/refclones.html
到目前(2015年7月31日15:17:59)已经有了205种,我爬取它们的genebank ID号,然后用python程序批量下载了它们的序列,能下载的序列共179条,都是8K左右的碱基序列。
根据genebank ID或者其它ID号批量下载核酸序列的脚本如下:
[python]</pre>
import sys
import time
import random
from Bio import Entrez
ids=[]
infile=sys.argv[1]
for line in open(infile,'r'):
line=line.strip()
ids.append(line)
for i in range(1,len(ids)):
# t = random.randrange(0,5)
handle =
Entrez.efetch(db="nucleotide", id=ids[i],rettype="fasta",email="jmzeng1314@163.com")
# time.sleep(t)
print handle.read()
[/python]
脚本使用很简单,保持输入文件是一行一个ID号即可。
同时,根据文献我们也能得到hbv病毒提取方法
当然,我是看不懂的。
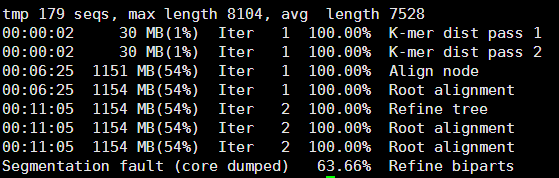
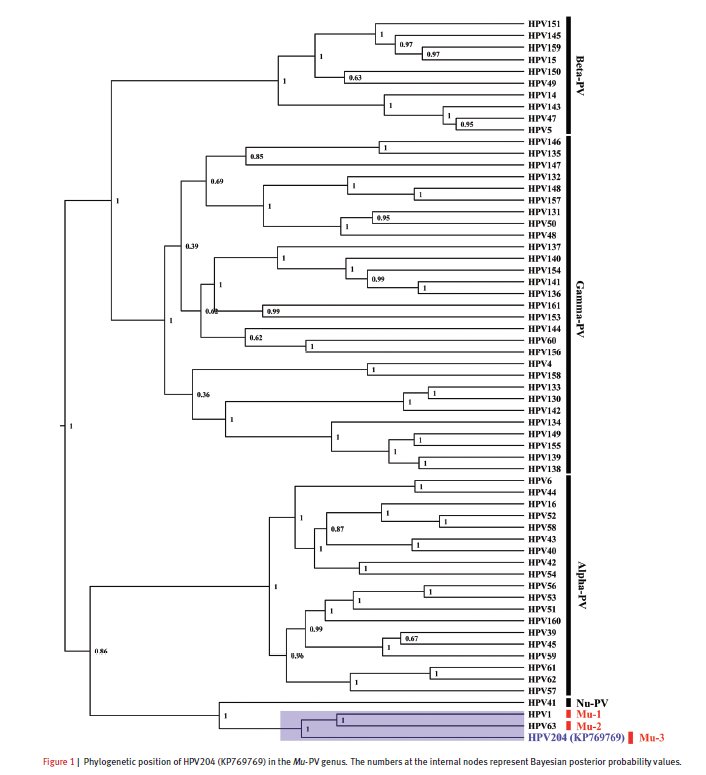
同样的拿到下载的178条序列我们可以做一个进化树,当然,这个文章已经做好了,我就不做了,进化树其实蛮简单的。
下载179条hpv序列,每条序列都是8KB左右
我还用了R脚本批量下载
library(ape)
a=read.table("hpv_all.ID") #输入文件是一行一个ID号即可
for (i in 1:nrow(a)){
tmp=read.GenBank(a[i,1],seq.names = a[1,1],as.character = T)
write.dna(tmp,"tmp.fa",format="fasta", append=T,colsep = "")
}
然后用muscle做比对,参照我之前的笔记
http://www.bio-info-trainee.com/?p=659
http://www.bio-info-trainee.com/?p=660
http://www.bio-info-trainee.com/?p=626
muscle -in mouse_J.pro -out mouse_J.pro.a
muscle -maketree -in mouse_J.pro.a -out mouse_J.phy
貌似时间有点长呀,最后还莫名其妙的挂掉了,可能是我的服务器配置有点低。
进化树如下所示: