Tag Archives: 癌症
对CCLE数据库可以做的分析
step4:Unsupervised hierarchical clustering (1-Spearman distance, average linkage) was performed on the cell lines using the aCGH data.
Putative driver genes of which copy number aberrations correlated to mRNA gene expression were identified to determine subtypes or clusters that are driven by different mechanisms. This was done using Mann Whitney U-test with p<0.05, and Spearman Correlation Coefficient test with Rho >0.6.
step5:We then performed consensus clustering[17] on the gene expression data of the 27 gastric cancer cell lines from CCLE using these putative driver genes. We selected k = 2 as it gives sufficiently stable similarity matrix.
step6: In order to assign new samples to this integrative cluster, significance analysis of microarray (SAM) [18]with threshold q<2.0 was used to generate subtype signature based on the mRNA expression data of the 1762 genes from the 27 gastric cancer cell lines in CCLE.
先用甲基化数据来聚类,得到putative driver genes,然后再用这些基因的表达数据来再次聚类,分成两类,然后对这两类进行SAM找差异基因
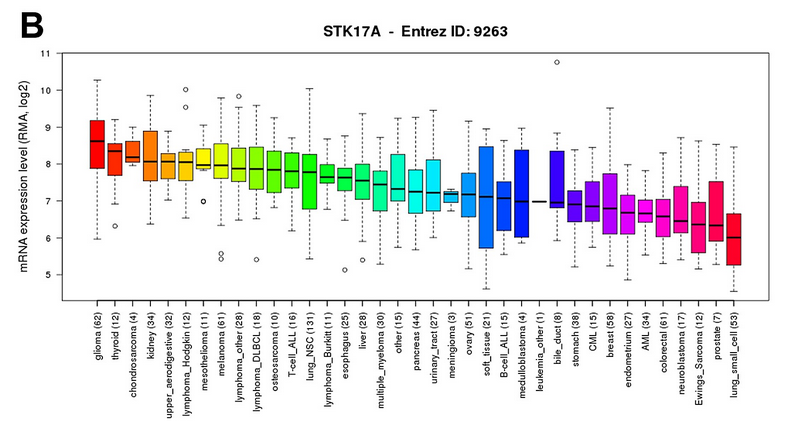
结论就是:STK17A is highly expressed in glioma cell lines compared to other cancer types. Data was obtained through the Cancer Cell Line Encyclopedia (CCLE).
第三篇文献:http://www.nature.com/ncomms/2013/130709/ncomms3126/fig_tab/ncomms3126_F4.html
寻找somatic突变的软件大合集
来自于:https://www.biostars.org/p/19104/
Here are a few more, a summary of the other answers, and updated links:
- deepSNV (abstract) (paper)
- EBCall (abstract) (paper)
- GATK SomaticIndelDetector (note: only available after an annoying sign-up and login)
- Isaac variant caller (abstract) (paper)
- joint-snv-mix (abstract) (paper)
- LoFreq (abstract) (paper) (call on tumor & normal separately and then use a filter to derive somatic events)
- MutationSeq (abstract) (paper)
- MutTect (abstract) (paper) (note: only available after an annoying sign-up and login)
- QuadGT (for calling single-nucleotide variants in four sequenced genomes comprising a normal-tumor pair and the two parents)
- samtools mpileup - by piping BCF format output from this to bcftools view and using the '-T pair' option
- Seurat (abstract) (paper)
- Shimmer (abstract) (paper)
- SolSNP (call on tumor & normal separately and then compare to identify somatic events)
- SNVMix (abstract) (paper)
- SOAPsnv
- SomaticCall (manual)
- SomaticSniper (abstract) (paper)
- Strelka (abstract) (paper)
- VarScan2 (abstract) (paper)
- Virmid (abstract) (paper)
For a much more general discussion of variant calling (not necessarily somatic or limited to SNVs/InDels) check out this thread: What Methods Do You Use For In/Del/Snp Calling?
Some papers describing comparisons of these callers:
- Comparing somatic mutation-callers: Beyond Venn diagrams.
- A comparative analysis of algorithms for somatic SNV detection in cancer
- Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers
- Comparison of somatic mutation calling methods in amplicon and whole exome sequence data.
The ICGC-TCGA DREAM Mutation Calling challenge has a component on somatic SNV calling.
This paper used validation data to compare popular somatic SNV callers:
Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers
You'll need to update the link to MuTect. Broad Institute has begun to put portable versions of their tools on Github, like thelatest release of MuTect. The Genome Institute at WashU has been using Github for a while, but portable versions of their tools can be found here and here.
To rehash/expand on what Dan said, if you're sequencing normal tissue, you generally expect to see single-nucleotide variant sites fall into one of three bins: 0%, 50%, or 100%, depending on whether they're heterozygous or homozygous.
With tumors, you have to deal with a whole host of other factors:
- Normal admixture in the tumor sample: lowers variant allele fraction (VAF)
- Tumor admixture in the normal - this occurs when adjacent normals are used, or in hematological cancers, when there is some blood in the skin normal sample
- Subclonal variants, which may occur in any fraction of the cells, meaning that your het-site VAF might be anywhere from 50% down to sub-1%, depending on the tumor's clonal architecture and the sensitivity of your method
- Copy number variants, cn-neutral loss of heterozygosity, or ploidy changes, all of which again shift the expected distribution of variant fractions
These, and other factors, make calling somatic variants difficult and still an area that is being heavily researched. If someone tells you that somatic variant calling is a solved problem, they probably have never tried to call somatic variants.
Sounds like somatic / tumor variant calling is something that will be solved by improvements at the wet lab side ( single cell selection / amplification / sequencing ) . Rather than at the computational side.
Well, single cell has a role to play (and would have more of one if WGA wasn't so lossy), but realistically, you can't sequence billions of cells from a tumor individually. Bulk sequencing still is going to have a role for quite a while.
Hell germ line calling isn't even a solved problem. Still get lots of false positives (and false negatives). It just tends to work so well that it is hard to improve it much except by making it faster, less memory intensive, etc
Solved was the wrong word. I just meant improved. There is only so much you can do at the computational side. Wet lab also has its part to play.
A germline variant caller generally has a ploidy-based genotyping algorithm built in to part of the algorithm/pipeline. I believe, IIRC, the GATK UnifiedGenotyper for instance does both variant calling and then genotype calling. So to call a genotype for a variant it is expecting a certain number of reads to support the alternative allele. When working with somatic variants all of the assumptions about how many reads you expect with a variant at a position to distinguish between true and false positives are no longer valid. Except for fixed mutations throughout the tumor population only some proportion of cells will hold a somatic variation. You also typically have some contamination from normal non-cancerous cells. Add in complications from significant genomic instability with lots of copy number variations and such and you have a need for a major change in your model for calling variation while minimizing artifactual calls. So you have a host of other programs that have been developed specifically for looking at somatic variation in tumor samples.
一篇文献:
Comparison of somatic mutation calling methods in amplicon and whole exome sequence data
是qiagen公司发的
High-throughput sequencing is rapidly becoming common practice in clinical diagnosis and cancer research. Many algorithms have been developed for somatic single nucleotide variant (SNV) detection in matched tumor-normal DNA sequencing. Although numerous studies have compared the performance of various algorithms on exome data, there has not yet been a systematic evaluation using PCR-enriched amplicon data with a range of variant allele fractions. The recently developed gold standard variant set for the reference individual NA12878 by the NIST-led “Genome in a Bottle” Consortium (NIST-GIAB) provides a good resource to evaluate admixtures with various SNV fractions.
Using the NIST-GIAB gold standard, we compared the performance of five popular somatic SNV calling algorithms (GATK UnifiedGenotyper followed by simple subtraction, MuTect, Strelka, SomaticSniper and VarScan2) for matched tumor-normal amplicon and exome sequencing data.
Nevertheless, detecting somatic mutations is still challenging, especially for low-allelic-fraction variants caused by tumor heterogeneity, copy number alteration, and sample degradation
We used QIAGEN’s GeneRead DNAseq Comprehensive Cancer Gene Panel (CCP, Version 1) for enrichment and library construction in triplicate。
QIAGEN’s GeneRead DNAseq Comprehensive Cancer Gene Panel (Version 1) was used to amplify the target region of interest (124 genes, 800 Kb).
When analyzing different types of data, use of different algorithms may be appropriate.
DNA samples of NA12878 and NA19129 were purchased from Coriell Institute. Sample mixtures were created based on the actual amplifiable DNA in each sample, resulting in 0%, 8%, 16%, 36%, and 100% of NA12878 sample mixed in the NA19129 sample, respectively.We treated the mixed samples at 8%, 16%, 36%, and 100% as the virtual tumor samples and the 0% as the virtual normal sample.
五个软件的算法是:
1. NaiveSubtract — SNVs were called separately from virtual tumor and normal samples using GATK UnifiedGenotyper [22]. For exome sequencing data, reads were already mapped, locally realigned and recalibrated by the 1,000 Genomes Project. So SNVs were directly called on the BAM files using GATK Unified Genotyper. Then, SNVs detected in the virtual normal sample were removed from the list of SNVs detected in the virtual tumor sample, leaving the “somatic” SNVs.
2. MuTect — MuTect is a method developed for detecting the most likely somatic point mutations in NGS data using a Bayesian classifier approach. The method includes pre-processing aligned reads separately in tumor and normal samples and post-processing resulting variants by applying an additional set of filters. We ran MuTect under the High-Confidence mode with its default parameter settings. We disabled the “Clustered position” filter and the “dbSNP filter” for the amplicon sequencing reads, and we disabled the “dbSNP filter” for the exome sequencing.
3. SomaticSniper — SomaticSniper calculates the Bayesian posterior probability of each possible joint genotype across the normal and cancer samples. We tuned the software’s parameters to increase sensitivity and then filtered raw results using a Somatic Score cut-off of 20 to improve specificity.
4. Strelka — Strelka reports the most likely genotype for tumor and normal samples based on a Bayesian probability model. Post-calling filters built into the software are based on factors such as read depth, mismatches, and overlap with indels. We skipped depth filtration for exome and amplicon sequencing data as recommended by the Strelka authors. For the amplicon sequencing reads, we set the minimum MAPQ score at 17 for consistency with the defaults in GATK UnifiedGenotyper. We used variants passing Strelka post-calling filters for analysis.
5. VarScan2 — VarScan2 performs analyses independently on pileup files from the tumor and normal samples to heuristically call a genotype at positions achieving certain thresholds of coverage and quality. Then, sites of the genotypes not matched in tumor and normal samples are classified into somatic, germline, or ambiguous groups using Fisher’s exact test. We generated the pileup files using SAMtools mpileup command.
The compatibility of the output VCF files between different methods as well as the NIST-GIAB gold standard was examined using bcbio.variation tools and manual inspection. The reported SNP call representations between files are comparable to each other.
使用oncotator做突变注释
做过癌症数据分析的童鞋都知道,TCGA里面用maf格式来记录突变!那么maf格式的数据是如何得来的呢,我们都知道,做完snp-calling一般是得到vcf格式的突变记录数据文件,然后再用annovar或者其它蛋白结构功能影响预测软件注释一下,还远达不到maf的近100条记录。
Genomic Annotations
- Gene, transcript, and functional consequence annotations using GENCODE for hg19.
- Reference sequence around a variant.
- GC content around a variant.
- Human DNA Repair Gene annotations from Wood et al.
Protein Annotations
Cancer Variant Annotations
- Observed cancer mutation frequency annotations from COSMIC.
- Cancer gene and mutation annotations from the Cancer GenCensus.
- Overlapping mutations from the Cancer Cell Line Encyclopedia.
- Cancer gene annotations from the Familial Cancer Database.
- Cancer variant annotations from ClinVar.
Non-Cancer Variant Annotations
- Common SNP annotations from dbSNP.
- Variant annotations from 1000 Genomes.
- Variant annotations from NHLBI GO Exome Sequencing Project (ESP).
2012-LAD的三个亚型的不同生物学意义
文献名:Differential Pathogenesis of Lung Adenocarcinoma Subtypes Involving Sequence Mutations, Copy Number, Chromosomal Instability, and Methylation
Lung adenocarcinoma (LAD)的遗传变异度很大。
这个癌症可以分成三类:The LAD molecular subtypes (Bronchioid, Magnoid, and Squamoid)
然后我们在三个subtypes里面分析了以下四个特征,发现不同subtypes差异非常显著。
1、Gene mutation rates (EGFR, KRAS, STK11, TP53),
2、chromosomal instability,
3、regional copy number
4、genomewide DNA methylation
另外三个临床特征也是很显著。
1、Patient overall survival,
2、cisplatin plus vinorelbine therapy response
3、predicted gefitinib sensitivity
所以,我们的分类非常好,而且对临床非常有帮助。
对LAD的研究数据包括
1,DNA copy number
2,gene sequence mutation
3,DNA methylation
4,gene expression
即使是TP53这样的基因在LAD的突变率也才35%,所以我们的LAD应该更加细分,因为EGFR mutation and KRAS mutation这样的突变对治疗很有指导意义,细分更加有助于临床针对性治疗方案的选择。
我们选取了116个LAD样本的数据,分析了1,genome-wide gene expression,,2,genomewide DNA copy number, 3,genome-wide DNA methylation, 4,selected gene sequence mutations
得到的结论是:LAD molecular subtypes correlate with grossly distinct genomic alterations and patient therapy response
数据来源如下:
Gene expression --> Agilent 44 K microarrays.
DNA copy number --> Affymetrix 250 K Sty and SNP6 microarrays.
DNA methylation --> MSNP microarray assay.
DNA from EGFR, KRAS, STK11 and TP53 exons --> ABI sequencers
我们用的是R语言包 ConsensusClusterPlus根据gene expression 来对我们的LAD进行分类molecular subtypes
分类的基因有506个(the top 25% most variable genes, 3,045, using ConsensusClusterPlus),A nearest centroid subtype predictor utilizing 506 genes
这三类LAD的过表达基因参与不同的生物功能,
Bronchioid – excretion genes, asthma genes, and surfactants (SFTPB, SFTPC, SFTPD);
Magnoid – DNA repair genes, such as thymine-DNA glycosylase (TDG);
Squamoid – defense response genes, such as chemokine ligand 10 (CXCL10)
而且也对应不同的临床数据
Bronchioid had the most females, nonsmokers, early stage tumors, and low grade tumors, the greatest acinar content, the least necrosis, and the least invasion.
Squamoid had the most high grade tumors, the greatest solid content, and the lowest papillary content.
Magnoid had themost smokers and the heaviest smokers by pack years.
它们的基因突变pattern也有很大区别。
Bronchioid had the greatest EGFR mutation frequency
Magnoid had the greatest mutation frequencies in TP53, KRAS and STK11.
为了研究不同亚型癌症的突变模式的不同(genomewide mutation rates),我们同时又研究了a large set of rarely mutated genes (n = 623) from the Ding et al. cohort
结论:
Bronchioid subtype 更有可能受益于EGFR inhibitory therapy
Magnoid tumors also have severe genomic alterations including the greatest CIN, the most regional CN alterations, DNA hypermethylation, and the greatest genomewide mutation rate.
the Squamoid subtype displayed the fewest distinctive alterations that included only regional CN alterations
2013-science-3205tumors-12types-4-ways-find-291HCD
2014-4742samples-21tumors-Cancer5000-set-254-genes
2015-MADGiC-identify-cancer-driver-gene
2014-REVIEW-identifying driver mutation in sequenced cancer genome
2014-review-Next-generation sequencing to guide cancer therapy
This reductionist thinking led the initial theories on carcinogenesis to be centered on how many “hits” or genetic mutations were necessary for a tumor to develop.
研究癌症领域必看文献
最近需要了解一些癌症相关知识,看到了这个文献列表,觉得非常棒,所以推荐给大家。
抽时间慢慢看,一个月应该可以把这些文献看完的。
癌症种类大全 http://www.cancer.gov/types
癌症药物大全 http://www.cancer.gov/about-cancer/treatment/drugs
癌症所有的信息几乎都能在这个网站上面找到 http://www.cancer.gov/
包括癌症的科普、treatment、diagnosis,prognosis,classification,drugs、prediction等等
Cancer Precision Medicine: Improving Evidence in Practice - August 24, 2015
NCI-MATCH Trial Opens,![]() AACR blog post, August 2015
AACR blog post, August 2015
NCI-MATCH launch highlights new trial design in precision-medicine era![]()
McNeal C , JNCI, August 2015
The Cancer Genomics Resource List, 2014![]()
Zutter MM et al. CAP Lab Improvement Program,Archives of Pathology, August 2015
Personalized medicine and economic evaluation in oncology: all theory and no practice?![]()
Garattini L et al. Expert Rev Pharmacoecon Outcomes Res 2015 Aug 9. 1-6
Precision medicine trials bring targeted treatments to more patients,![]() C. Helwick, ASCO Post, Jul 25
C. Helwick, ASCO Post, Jul 25
Next-generation sequencing to guide cancer therapy ![]()
Gagan J et al, Genome Medicine, July 29, 2015
Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials.![]()
Meric-Bernstam F et al. J. Clin. Oncol. 2015 May 26.
Brave-ish new world-what's needed to make precision oncology a practical reality.![]()
MacConaill LE et al. JAMA Oncol 2015 Jul 16.
Genomic profiling: Building a continuum from knowledge to care![]()
Helen C et al. JAMA Oncology, July 2015
Are we there yet?![]()
When it comes to curing cancer, targeted therapies and genomic sequencing are helping, but we still have far to go. Genome Magazine, June 29, 2015
Artificial intelligence, big data, and cancer![]()
Kantarjian H et al, JAMA Oncology, June 2015
Multigene panel testing in oncology practice - how should we respond?![]()
Kurian AW et al. JAMA Oncology, June 2015
Use of whole genome sequencing for diagnosis and discovery in the cancer genetics clinic.![]()
Foley SB et al. EBioMedicine 2015 Jan 2(1) 74-81
The future of molecular medicine: biomarkers, BATTLEs, and big data ![]()
ES Kim, ASCO University, June 2015
NCI-MATCH trial will link targeted cancer drugs to gene abnormalities![]()
Targeted agent and profiling utilization registry study,![]() from the American Society for Clinical Oncology
from the American Society for Clinical Oncology
ASCO study aims to learn from patient access to targeted cancer drugs used off-label,![]() American Society for Clinical Oncology
American Society for Clinical Oncology
Improving evidence developed from population-level experience with targeted agents ![]() [PDF 462.93 KB]
[PDF 462.93 KB]![]()
McLellan M et al Issue Brief. Conference on Clinical Cancer Research November 2014
Implementing personalized cancer care.![]()
Schilsky RL et al. Nat Rev Clin Oncol 2014 Jul (7) 432-8
Accelerating the delivery of patient-centered, high-quality cancer care.![]()
Abrahams E et al. Clin. Cancer Res. 2015 May 15. (10) 2263-7
Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity.![]()
Catenacci DV et al. Mol Oncol 2015 May (5) 967-996
Cancer Precision Medicine: Improving Evidence in Practice - May 29, 2015
Diagnosis and treatment of cancer using genomics![]()
Vockley JG et al. BMJ, May 28, 2015
Targeted agent and profiling utilization registry study,![]() from the American Society for Clinical Oncology
from the American Society for Clinical Oncology
ASCO study aims to learn from patient access to targeted cancer drugs used off-label,![]() American Society for Clinical Oncology
American Society for Clinical Oncology
Improving evidence developed from population-level experience with targeted agents ![]() [PDF 462.93 KB]
[PDF 462.93 KB]![]()
McLellan M et al Issue Brief. Conference on Clinical Cancer Research November 2014
Implementing personalized cancer care.![]()
Schilsky RL et al. Nat Rev Clin Oncol 2014 Jul (7) 432-8
Accelerating the delivery of patient-centered, high-quality cancer care.![]()
Abrahams E et al. Clin. Cancer Res. 2015 May 15. (10) 2263-7
Next-generation clinical trials: Novel strategies to address the challenge of tumor molecular heterogeneity.![]()
Catenacci DV et al. Mol Oncol 2015 May (5) 967-996
Precision Medicine: Cancer and Genomics - May 12, 2015
Promise, peril seen in personalized cancer therapy,![]() by Marie McCullough, Philadelphia Inquirer, May 10
by Marie McCullough, Philadelphia Inquirer, May 10
A decision support framework for genomically informed investigational cancer therapy.![]()
Meric-Bernstam F et al. J. Natl. Cancer Inst. 2015 Jul (7)
Divide and conquer: The molecular diagnosis of cancer,![]() by Louis M. Staudt, National Cancer Insitute, Apr 13
by Louis M. Staudt, National Cancer Insitute, Apr 13
Health: Make precision medicine work for cancer care![]()
To get targeted treatments to more cancer patients pair genomic data with clinical data, and make the information widely accessible, Mark A. Rubin. Nature News, Apr 15
Using somatic mutations to guide treatment decisions![]()
Horlings H et al. JAMA Oncology, March 12, 2015
The landscape of precision cancer medicine clinical trials in the United States![]()
Roper N et al. Cancer Treatment Reviews 2015
What is “precision medicine?![]() Information from the National Cancer Institute
Information from the National Cancer Institute
Impact of cancer genomics on precision medicine for the treatment of cancer,![]() from the Cancer Genome Atlas, NCI
from the Cancer Genome Atlas, NCI
US precision-medicine proposal sparks questions,![]() by Sara Reardon, Nature News, Jan 22
by Sara Reardon, Nature News, Jan 22
Obama's 'precision medicine' means gene mapping,![]() NBC News, Jan 21
NBC News, Jan 21
What is President Obama's 'precision medicine' plan, and how might it help you?![]() By Lenny Bernstein, Jan 21
By Lenny Bernstein, Jan 21
Recent reviews
Companion diagnostics: the key to personalized medicine.![]()
Jørgensen JT. Expert Rev Mol Diagn. 2015 Feb;15(2):153-6
Promoting precision cancer medicine through a community-driven knowledgebase.![]()
Geifman N, et al. J Pers Med. 2014 Dec 15;4(4):475-88.
Toward a prostate cancer precision medicine.![]()
Rubin MA. Urol Oncol. 2014 Nov 20.
Prioritizing targets for precision cancer medicine.![]()
Andre F, et al. Ann Oncol. 2014 Dec;25(12):2295-303
Toward precision medicine with next-generation EGFR inhibitors in non-small-cell lung cancer.![]()
Yap TA, Popat S. Pharmgenomics Pers Med. 2014 Sep 19;7:285-95.
Genomically driven precision medicine to improve outcomes in anaplastic thyroid cancer.![]()
Pinto N, et al. J Oncol. 2014;936285
Translating genomics for precision cancer medicine.![]()
Roychowdhury S, Chinnaiyan AM. Annu Rev Genomics Hum Genet. 2014;15:395-415
The Cancer Genome Atlas: Accomplishments and Future - April 3, 2015
The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge![]()
Tomczak K, et al. Contemp Oncol (Pozn). 2015; 19(1A): A68-A77.
The Cancer Genome Atlas' 4th Annual Scientific Symposium![]()
May 11-12 ~ Bethesda, MD
The Cancer Genome Atlas (TCGA) Data Portal ![]()
Portal provides a platform for researchers to search, download, and analyze data sets generated by TCGA
Cancer Genomics Hub: A resource of the National Cancer Institute,![]() from the USC Genome Browser
from the USC Genome Browser
Molecular classification of gastric adenocarcinoma: translating new insights from The Cancer Genome Atlas Research Network.![]()
Sunakawa Y et al. Curr Treat Options Oncol 2015 Apr (4) 331
TCGA data and patient-derived orthotopic xenografts highlight pancreatic cancer-associated angiogenesis.![]()
Gore J et al. Oncotarget 2015 Feb 25.
Radiogenomics of clear cell renal cell carcinoma: preliminary findings of The Cancer Genome Atlas-Renal Cell Carcinoma (TCGA-RCC) Imaging Research Group.![]()
Shinagare AB et al. Abdom Imaging 2015 Mar 10.
Proteomics of colorectal cancer in a genomic context: First large-scale mass spectrometry-based analysis from the Cancer Genome Atlas.![]()
Jimenez CR et al. Clin. Chem. 2015 Feb 26.
End of cancer-genome project prompts rethink![]()
Geneticists debate whether focus should shift from sequencing genomes to analysing function. Heidi Ledford, Nature News and Comments, January 2015
Cancer Genomics: Insights into Driver Mutations - March 10, 2015
Seek and destroy: Relating cancer drivers to therapies![]()
E. Martinez-Ledesma et al. Cell, March 9, 2015
In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities![]()
C Rubio-Perez et al. Cancer Cell, March 9, 2015
MADGiC: a model-based approach for identifying driver genes in cancer. ![]() [PDF 373.56 KB]
[PDF 373.56 KB]![]()
Keegan D. Korthauer et al. Bioinformatics, January 2015
Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine.![]()
Benjamin J Raphael et al. Genome Medicine 2014
Novel recurrently mutated genes in African American colon cancers.![]()
Guda K et al. Proc Natl Acad Sci U S A. 2015 Jan 12
Sparse expression bases in cancer reveal tumor drivers.![]()
Logsdon BA, et al. Nucleic Acids Res. 2015 Jan 12
Patient-specific driver gene prediction and risk assessment through integrated network analysis of cancer omics profiles.![]()
Bertrand D, et al. Nucleic Acids Res. 2015 Jan 8
Identification of constrained cancer driver genes based on mutation timing.![]()
Sakoparnig T, et al. PLoS Comput Biol. 2015 Jan 8;11(1):e1004027
CaMoDi: a new method for cancer module discovery.![]()
Manolakos A, et al. BMC Genomics. 2014 Dec 12;15 Suppl 10:S8.
VHL, the story of a tumour suppressor gene.![]()
Gossage L, et al. Nat Rev Cancer. 2014 Dec 23;15(1):55-64
Targeting the MET pathway for potential treatment of NSCLC.![]()
Li A, et al. Expert Opin Ther Targets. 2014 Dec 23:1-12
Deciphering oncogenic drivers: from single genes to integrated pathways.![]()
Chen J, et al. Brief Bioinform. 2014 Nov 5.
Driver and passenger mutations in cancer.![]()
Pon JR, et al. Annu Rev Pathol. 2014 Oct 17
Hereditary Cancer Genetic Testing: Where are We? - December 18, 2014
NCI paper:Prevalence and correlates of receiving and sharing high-penetrance cancer genetic test results: Findings from the Health Information National Trends Survey![]()
Taber J.M. et al Public Health Genomics, January 2015
Clinical decisions: Screening an asymptomatic person for genetic risk--polling results![]()
Schulte J, et al. N Engl J Med 2014 Nov;371(20):e30
Testing for hereditary breast cancer: Panel or targeted testing? Experience from a clinical cancer genetics practice.![]()
Doherty J, J Genet Couns. 2014 Dec 5
Hereditary colorectal cancer syndromes: American Society of Clinical Oncology clinical practice guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology clinical practice guidelines.![]()
Stoffel EM, et al. J Clin Oncol. 2014 Dec 1
Population testing for cancer predisposing BRCA1/BRCA2 mutations in the Ashkenazi-Jewish community: A randomized controlled trial.![]()
Manchanda R, et al. J Natl Cancer Inst. 2014 Nov 30;107(1)
Cost-effectiveness of population screening for BRCA mutations in Ashkenazi Jewish women compared with family history-based testing.![]()
Manchanda R et al. J Natl Cancer Inst. 2014 Nov 30;107(1). pii: dju380. doi: 10.1093/jnci/dju380. Print 2015 Jan.
Check out our Cancer Genetic Testing Update Page for additional information and links
Cancer Genomic Tests (October 30, 2014)
Cancer Genomic Tests: Accelerating Translation - October 30, 2014
CDC-NCI paper: An overview of recommendations and translational milestones for genomic tests in cancer![]()
Christine Q. Chang et al. Genetics in Medicine, October 22, 2014
Check out the CDC evidence-based classification of cancer genomic tests
Check out the NCI Cancer Genomics and Epidemiology Navigator for latest information on cancer genomic tests![]()
EGAPP: A model process for evaluating genomic applications in practice and prevention.![]() Check out cancer genomic tests, methods, evidence reviews and recommendation statements.
Check out cancer genomic tests, methods, evidence reviews and recommendation statements.
NCI Fact Sheet: Genetic testing for hereditary cancer syndromes![]()
Cancer Genomics: Impact of Recent Insights - October 30, 2014
Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin.![]()
Katherine A. Hoadley et al. Cell, August 2014
Genome study overhauls cancer categories, shifts from tissues to molecular subtypes,![]() by Kevin Mayer, Genetics Bioengineering News, Aug 8
by Kevin Mayer, Genetics Bioengineering News, Aug 8
It's time for us to think about cancer differently,![]() by Paula Mejia, Newsweek, Aug 8
by Paula Mejia, Newsweek, Aug 8
NIH- The Cancer Genome Atlas (TCGA) Initiative![]()
NIH information: What is cancer genomics and the genetic basis of cancer?![]()
Cancer Precision Medicine: Where Are We? - September 18, 2014
NIH announces the launch of 3 integrated precision medicine trials; ALCHEMIST is for patients with certain types of early-stage lung cancer,![]() August 2014
August 2014
National Cancer Institute's Precision Medicine Initiatives for the New National Clinical Trials Network.![]() Jeffrey Abrams et al. ASCO Annual Meeting 2014
Jeffrey Abrams et al. ASCO Annual Meeting 2014
Personalized medicine: Special treatment.![]()
Michael Eisenstein. Nature, September 11, 2014
Why the controversy? Start sequencing tumor genes at diagnosis. Tumor sequencing at the time of diagnosis can give significant insight for successful cancer treatment,![]() by Shelly Gunn, Genetic Engineering & Biotechnology News, Sep 10
by Shelly Gunn, Genetic Engineering & Biotechnology News, Sep 10
National Cancer Institute information: Precision medicine and targeted therapy![]()
Genomics and precision oncology: What's a targeted therapy for cancer?![]() An updated list of approved drugs from the National Cancer Institute (2014)
An updated list of approved drugs from the National Cancer Institute (2014)
Therapy: This time it's personal![]()
Gravitz L Nature 509, S52-S54 2014 May 29
Multi-marker solid tumor panels using next-generation sequencing to direct molecularly targeted therapies![]()
Michael Marrone, et al. PLoS Currents Evidence on Genomic Tests 2014 May 27
Impact of cancer genomics on precision medicine for the treatment of cancer,![]() from the National Cancer Institute
from the National Cancer Institute
Cancer genomics and precision medicine in the 21st century ![]() [PDF 2.20 MB]
[PDF 2.20 MB]![]() , power point presentation from the National Human Genome Research Institute
, power point presentation from the National Human Genome Research Institute