mkdir -p ~/annotation/variation/human/dbSNPcd ~/annotation/variation/human/dbSNP## https://www.ncbi.nlm.nih.gov/projects/SNP/## ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606_b147_GRCh38p2/## ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606_b147_GRCh37p13/nohup wget ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606_b147_GRCh37p13/VCF/All_20160601.vcf.gz &wget ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606_b147_GRCh37p13/VCF/All_20160601.vcf.gz.tbi
Tag Archives: GWAS
GWAS研究现状及资源下载
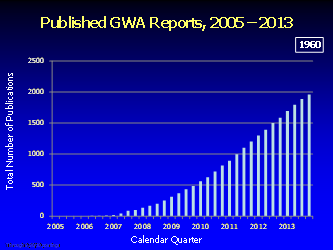
GWAS研究是非常火的,NHGIR还专门为它开辟了专栏来介绍,下面这个图片也是来自于NHGIR组织,是GWAS近年来发表文章的状况。
可以在该文章上面下载这个所有的数据
wget http://www.genome.gov/admin/gwascatalog.txt
截至目前为止。2015年5月8日21:08:34
这个文档有19603行的数据,但是只有2113篇pubmed文献,共涉及到七千多个基因
有293种杂志都发过GWAS的文章,总共有2113篇文献,发表关联分析突变位点最多的是这篇文献23251661 在PLoS One杂志上面,共 949个rs突变
杂志排序
cut -f 2,5 gwascatalog.txt |perl -alne '{$hash{$_}++}END{print "$_" foreach sort {$hash{$a} <=> $hash{$b}} keys %hash}' |cut -f 2 |perl -alne '{$hash{$_}++}END{print "$_\t$hash{$_}" foreach sort {$hash{$a} <=> $hash{$b}} keys %hash}'
Hum Genet 41
Am J Hum Genet 62
Mol Psychiatry 64
PLoS One 132
PLoS Genet 145
Hum Mol Genet 168
Nat Genet 397
文章的rs突变点排序
cut -f 2,5 gwascatalog.txt |perl -alne '{$hash{$_}++}END{print "$_ $hash{$_}" foreach sort {$hash{$a} <=> $hash{$b}} keys %hash}'
24324551 PLoS One 241
24097068 Nat Genet 245
24816252 Nat Genet 299
23382691 PLoS Genet 699
23251661 PLoS One 949
数据打开如下:
我取了表头和第一行数据,然后把它转置了,这样方便查看
| Date Added to Catalog | 10/22/2014 |
| PUBMEDID | 24528284 |
| First Author | Ji Y |
| Date | 08/01/2014 |
| Journal | Br J Clin Pharmacol |
| Link | http://www.ncbi.nlm.nih.gov/pubmed/24528284 |
| Study | Citalopram and escitalopram plasma drug and metabolite concentrations: genome-wide associations. |
| Disease/Trait | Response to serotonin reuptake inhibitors in major depressive disorder (plasma drug and metabolite levels) |
| Initial Sample Description | 300 European ancestry Escitalpram treated individuals, 130 European ancestry Citalopram treated individuals |
| Replication Sample Description | NA |
| Region | 17q25.3 |
| Chr_id | 17 |
| Chr_pos | 79831041 |
| Reported Gene(s) | CBX4 |
| Mapped_gene | CBX8 - CBX4 |
| Upstream_gene_id | 57332 |
| Downstream_gene_id | 8535 |
| Snp_gene_ids | |
| Upstream_gene_distance | 33.93 |
| Downstream_gene_distance | 2.12 |
| Strongest SNP-Risk Allele | rs9747992-? |
| SNPs | rs9747992 |
| Merged | 0 |
| Snp_id_current | 9747992 |
| Context | Intergenic |
| Intergenic | 1 |
| Risk Allele Frequency | 0.086 |
| p-Value | 2.00E-07 |
| Pvalue_mlog | 6.698970004 |
| p-Value (text) | (S-DCT concentration) |
| OR or beta | NR |
| 95% CI (text) | NR |
| Platform [SNPs passing QC] | Illumina [7,537,437] (Imputed) |
| CNV | N |
上面这个文件是由tab键分割的,每一列的意义如下!
Note: The SNP data in the catalog has been mapped to dbSNP Build 142 and Genome Assembly,
GRCh38/hg37.p13.
DATE ADDED TO CATALOG: Date added to catalog
PUBMEDID: PubMed identification number
FIRST AUTHOR: Last name of first author
DATE: Publication date (online (epub) date if available)
JOURNAL: Abbreviated journal name
LINK: PubMed URL
STUDY: Title of paper (linked to PubMed abstract)
DISEASE/TRAIT: Disease or trait examined in study
INITIAL SAMPLE SIZE: Sample size for Stage 1 of GWAS
REPLICATION SAMPLE SIZE: Sample size for subsequent replication(s)
REGION: Cytogenetic region associated with rs number (NCBI)
CHR_ID: Chromosome number associated with rs number (NCBI)
CHR_POS: Chromosomal position associated with rs number (dbSNP Build 132,
NCBI)
REPORTED GENE (S): Gene(s) reported by author
MAPPED GENE(S): Gene(s) mapped to the strongest SNP (NCBI). If the SNP is
located within a gene, that gene is listed. If the SNP is intergenic, the upstream and
downstream genes are listed, separated by a hyphen. UPSTREAM_GENE_ID:
Entrez Gene ID for nearest upstream gene to rs number, if not within gene (NCBI)
DOWNSTREAM_GENE_ID: Entrez Gene ID for nearest downstream gene to rs
number, if not within gene (NCBI)
SNP_GENE_IDS: Entrez Gene ID, if rs number within gene; multiple genes
denotes overlapping transcripts (NCBI)
UPSTREAM_GENE_DISTANCE: distance in kb for nearest upstream gene to rs
number, if not within gene (NCBI)
DOWNSTREAM_GENE_DISTANCE: distance in kb for nearest downstream
gene to rs number, if not within gene (NCBI)
STRONGEST SNP-RISK ALLELE: SNP(s) most strongly associated with trait +
risk allele (? for unknown risk allele). May also refer to a haplotype.
SNPS: Strongest SNP; if a haplotype is reported above, may include more than one
rs number (multiple SNPs comprising the haplotype)
MERGED: denotes whether the SNP has been merged into a subsequent rs record
(0 = no; 1 = yes; NCBI)
SNP_ID_CURRENT: current rs number (will differ from strongest SNP when
merged = 1)
CONTEXT: SNP functional class (NCBI)
INTERGENIC: denotes whether SNP is in intergenic region (0 = no; 1 = yes;
NCBI)
RISK ALLELE FREQUENCY: Reported risk allele frequency associated with
strongest SNP
P-VALUE: Reported p-value for strongest SNP risk allele (linked to dbGaP
Association Browser)
PVALUE_MLOG: -log(p-value)
P-VALUE (TEXT): Information describing context of p-value (e.g. females,
smokers).
Note that p-values are rounded to 1 significant digit (for example, a published pvalue of 4.8 x 10-7 is rounded to 5 x 10-7).
OR or BETA: Reported odds ratio or beta-coefficient associated with strongest
SNP risk allele
95% CI (TEXT): Reported 95% confidence interval associated with strongest SNP
risk allele
PLATFORM (SNPS PASSING QC): Genotyping platform manufacturer used in
Stage 1; also includes notation of pooled DNA study design or imputation of
SNPs, where applicable
CNV: Study of copy number variation (yes/no)
Updated: January 13, 2015