Tag Archives: TCGA
TCGA表达数据的多项应用之4–求指定基因在指定癌症里面的表达量相关性矩阵,与所有的基因比较。
这个不出图,会给出TCGA里面涉及到的所有基因跟你指定的基因的表达量相关系数和P值,分别你一次性的看清楚你感兴趣的基因跟体内其它基因在该癌症种类的相关性,当然,相关非因果,请谨慎应用! Continue reading
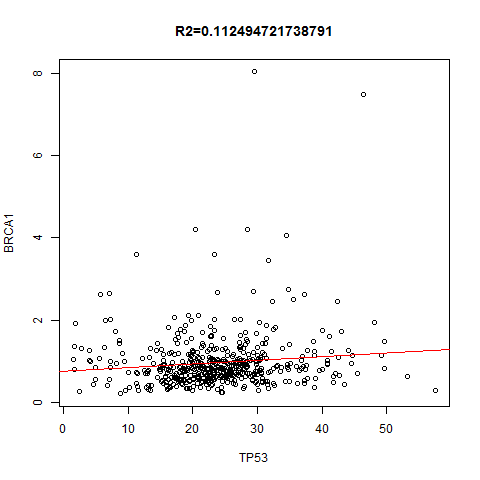
TCGA表达数据的多项应用之3–对指定的两个基因,在所有癌种里面找到correlation并作图
上面是指定一个基因在不同的癌种里面,本次讲指定任意两个基因,在所有癌种里面找到correlation并作图!图如下:
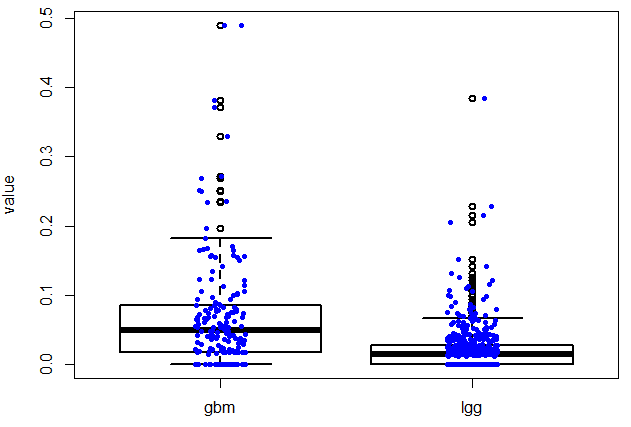
TCGA表达数据的多项应用之2–对指定基因在不同癌种里面画boxplot,或者在所有的normal组织里面看表达量!
好像文章题目没有长度限制,太好了!本讲所实现的目标非常简单,如题,指定基因在不同癌种里面画boxplot,或者在所有的normal组织里面看表达量!下面是一个具体的例子:
TCGA表达数据的多项应用之1–下载数据并且导入mysql
2016-TCGA数据挖掘系列文章之癌症男女有别
TCGA数据挖掘系列文章之-pseudogene假基因探究
所有TCGA的maf格式somatic突变数据均可下载
如果你研究癌症,那么TCGA计划的如此丰富的公共数据你肯定不能错过,一般人只能获取到level3的数据,当然,其实一般人也没办法使用level1和level2的数据,毕竟近万个癌症样本的原始测序数据,还是很恐怖的,而且我们拿到原始数据,再重新跑pipeline,其实并不一定比人家TCGA本身分析的要好,所以我们直接拿到分析结果,就足够啦!
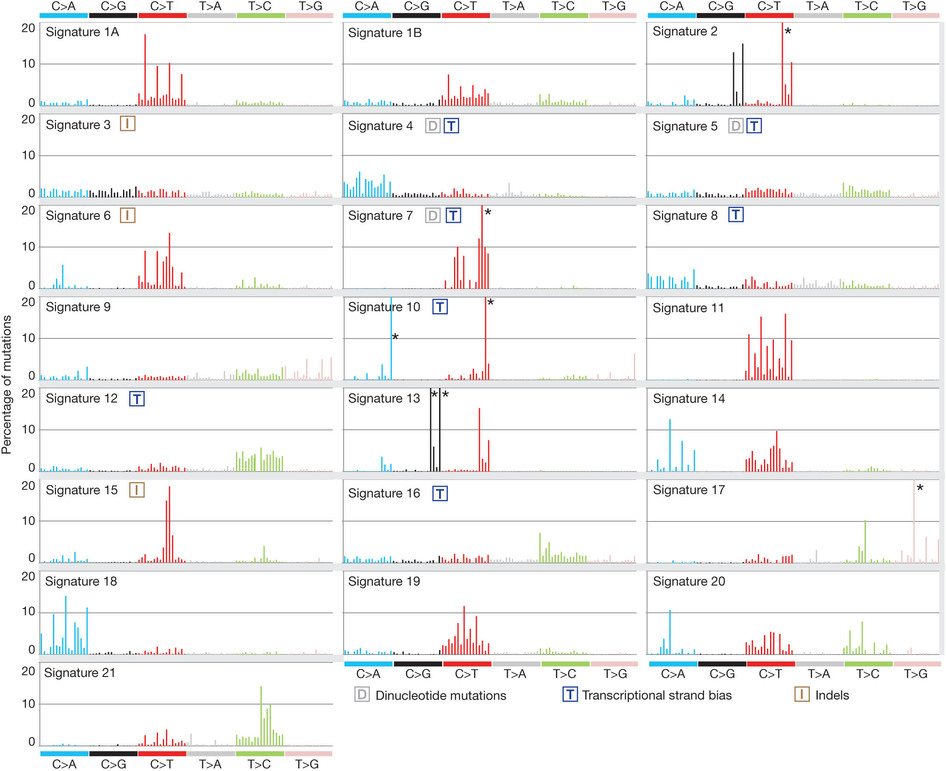
突变频谱探究mutation siganures
这也是对TCGA数据的深度挖掘,从而提出的一个统计学概念。文章研究了30种癌症,发现21种不同的mutation signature。如果理解了,就会发现这个其实蛮简单的,他们并不重新测序,只是拿已经有了的TCGA数据进行分析,而且居然是发表在nature上面!
研究了4,938,362 mutations from 7,042 cancers样本,突变频谱的概念只是针对于somatic 的mutation。一般是对癌症病人的肿瘤组织和癌旁组织配对测序,过滤得到的somatic mutation,一般一个样本也就几百个somatic 的mutation。
paper链接是:http://www.nature.com/nature/journal/v500/n7463/full/nature12477.html
用TCGA数据做cox生存分析的风险因子(比例风险模型)
用my.surv <- surv(OS_MONTHS,OS_STATUS=='DECEASED')构建生存曲线。用kmfit2 <- survfit(my.surv~TUMOR_STAGE_2009)来做某一个因子的KM生存曲线。用 survdiff(my.surv~type, data=dat)来看看这个因子的不同水平是否有显著差异,其中默认用是的logrank test 方法。用coxph(Surv(time, status) ~ ph.ecog + tt(age), data=lung) 来检测自己感兴趣的因子是否受其它因子(age,gender等等)的影响。
TCGA数据里面的生存分析例子
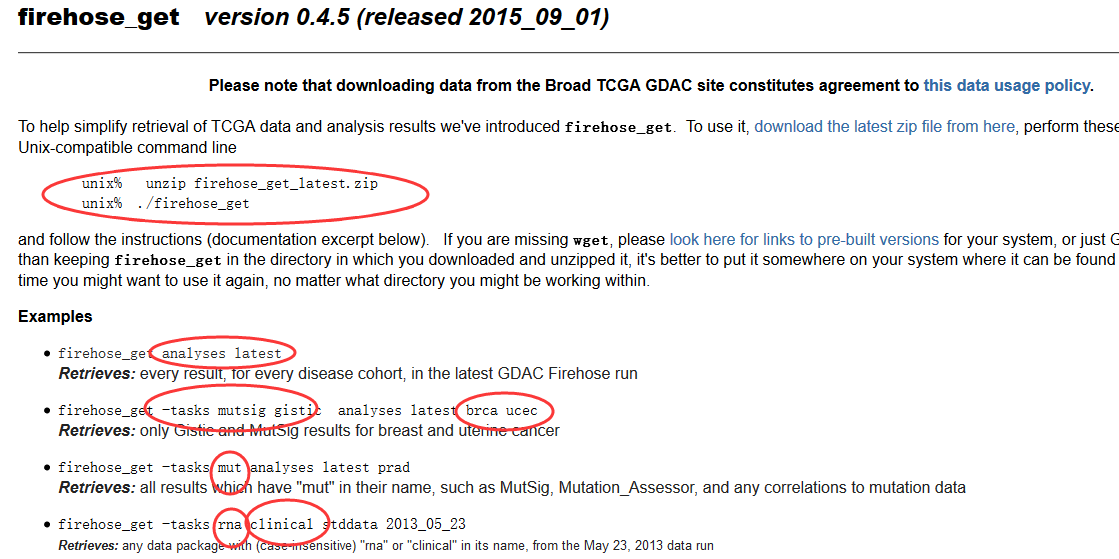
用firehose_get 来下载所有TCGA寄存在broad的数据
ACC BLCA BRCA CESC COAD COADREAD DLBC ESCA GBM HNSC KICH KIRC KIRP LAML LGG LIHC LUAD LUSC OV PAAD PANCANCER PANCAN8 PANCAN12 PRAD READ SARC SKCM STAD THCA UCEC UCS
做癌症研究一定要把这几十篇TCGA的大文章看完
使用R包cgdsr来下载TCGA的数据
前面我讲到TCGA的数据可以在5个组织机构可以获取,他们都提供了类似的接口来供用户下载数据
每个接口都有使用教程,比如http://firebrowse.org/tutorial/FireBrowse-Tutorial.pdf
非常详细!!!
有的还专门写了软件接口:https://confluence.broadinstitute.org/display/GDAC/Download
或者写了R的接口:http://www.cbioportal.org/cgds_r.jsp
接下来我们要讲的就是cbioportal网站提供的一个R接口,非常好用,只需记住4个函数即可!!! Continue reading
TCGA数据下载大全
The molecular taxonomy of primary prostate cancer
Cell Volume 163 Issue 4: p1011-1025 Read the full article
Portal Publication Site and Associated Data Files
Comprehensive Molecular Characterization of Papillary Renal Cell Carcinoma
NEJM. Published on line on Nov 4th, 2015 Read the full article
Portal Publication Site and Associated Data Files
Tools for Exploring Data and Analyses
- Broad Institute FireBrowse portal, The Broad Institute
- cBioPortal for Cancer Genomics, Memorial Sloan-Kettering Cancer Center
- TCGA Batch Effects, MD Anderson Cancer Center
- Regulome Explorer, Institute for Systems Biology
- Next-Generation Clustered Heat Maps, MD Anderson Cancer Center
TCGA Data Portal
Data Levels and Data Types
broad_institute收集的癌症数据
| 肾上腺皮质 | Adrenocortical carcinoma | ACC | 92 | Browse | Browse |
| 膀胱,尿路上皮 | Bladder urothelial carcinoma | BLCA | 412 | Browse | Browse |
| 乳腺癌 | Breast invasive carcinoma | BRCA | 1098 | Browse | Browse |
| 子宫颈 | Cervical and endocervical cancers | CESC | 307 | Browse | Browse |
| 胆管癌 | Cholangiocarcinoma | CHOL | 36 | Browse | Browse |
| 结肠腺癌 | Colon adenocarcinoma | COAD | 460 | Browse | Browse |
| 大肠腺癌 | Colorectal adenocarcinoma | COADREAD | 631 | Browse | Browse |
| 淋巴肿瘤弥漫性大B细胞淋巴瘤 | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma | DLBC | 58 | Browse | Browse |
| 食管 | Esophageal carcinoma | ESCA | 185 | Browse | Browse |
| FFPE试点二期 | FFPE Pilot Phase II | FPPP | 38 | None | Browse |
| 胶质母细胞瘤 | Glioblastoma multiforme | GBM | 613 | Browse | Browse |
| 脑胶质瘤 | Glioma | GBMLGG | 1129 | Browse | Browse |
| 头颈部鳞状细胞癌 | Head and Neck squamous cell carcinoma | HNSC | 528 | Browse | Browse |
| 肾嫌色 | Kidney Chromophobe | KICH | 113 | Browse | Browse |
| 泛肾 | Pan-kidney cohort (KICH+KIRC+KIRP) | KIPAN | 973 | Browse | Browse |
| 肾透明细胞癌 | Kidney renal clear cell carcinoma | KIRC | 537 | Browse | Browse |
| 肾乳头细胞癌 | Kidney renal papillary cell carcinoma | KIRP | 323 | Browse | Browse |
| 急性髓系白血病 | Acute Myeloid Leukemia | LAML | 200 | Browse | Browse |
| 脑低级神经胶质瘤 | Brain Lower Grade Glioma | LGG | 516 | Browse | Browse |
| 肝癌 | Liver hepatocellular carcinoma | LIHC | 377 | Browse | Browse |
| 肺腺癌 | Lung adenocarcinoma | LUAD | 585 | Browse | Browse |
| 肺鳞状细胞癌 | Lung squamous cell carcinoma | LUSC | 504 | Browse | Browse |
| 间皮瘤 | Mesothelioma | MESO | 87 | Browse | Browse |
| 卵巢浆液性囊腺癌 | Ovarian serous cystadenocarcinoma | OV | 602 | Browse | Browse |
| 胰腺癌 | Pancreatic adenocarcinoma | PAAD | 185 | Browse | Browse |
| 嗜铬细胞瘤和副神经节瘤 | Pheochromocytoma and Paraganglioma | PCPG | 179 | Browse | Browse |
| 前列腺癌 | Prostate adenocarcinoma | PRAD | 499 | Browse | Browse |
| 直肠腺癌 | Rectum adenocarcinoma | READ | 171 | Browse | Browse |
| 肉瘤 | Sarcoma | SARC | 260 | Browse | Browse |
| 皮肤皮肤黑色素瘤 | Skin Cutaneous Melanoma | SKCM | 470 | Browse | Browse |
| 胃腺癌 | Stomach adenocarcinoma | STAD | 443 | Browse | Browse |
| 胃和食管癌 | Stomach and Esophageal carcinoma | STES | 628 | Browse | Browse |
| 睾丸生殖细胞肿瘤 | Testicular Germ Cell Tumors | TGCT | 150 | Browse | Browse |
| 甲状腺癌 | Thyroid carcinoma | THCA | 503 | Browse | Browse |
| 胸腺瘤 | Thymoma | THYM | 124 | Browse | Browse |
| 子宫内膜癌 | Uterine Corpus Endometrial Carcinoma | UCEC | 560 | Browse | Browse |
| 子宫癌肉瘤 | Uterine Carcinosarcoma | UCS | 57 | Browse | Browse |
| 葡萄膜黑色素瘤 | Uveal Melanoma | UVM | 80 | Browse | Browse |
看起来癌症很多呀,任重道远
TCGA数据库的癌症种类以及癌症相关基因列表
TCGA projects 里面包含的癌症种类非常多,但是我们分析数据时候常常用pan-cancer 12,pan-cancer 17,pan-cancer 21来表示数据集有多少种癌症,一般文献会给出癌症的简称或者全名:
BLCA, BRCA, COADREAD, GBM, HNSC, KIRC, LAML, LGG, LUAD, LUSC, OV, PRAD, SKCM, STAD, THCA, UCEC.
Acute myeloid leukaemia
Bladder
Breast
Carcinoid
Chronic lymphocytic leukaemia
Colorectal
Diffuse large B-cell lymphoma
Endometrial
Oesophageal adenocarcinoma
Glioblastoma multiforme
Head and neck
Kidney clear cell
Lung adenocarcinoma
Lung squamous cell carcinoma
Medulloblastoma
Melanoma
Multiple myeloma
Neuroblastoma
Ovarian
Prostate
Rhabdoid tumour
HCD features: download
这是高置信度的癌症驱动基因列表:共280多个基因
Cancer5000 features: download
这是一篇对接近5000个癌症样本的研究得到的癌症相关基因列表:共230多个基因
参考:http://bg.upf.edu/oncodrive-role/
http://bioinformatics.oxfordjournals.org/content/30/17/i549.full
http://www.nature.com/nature/journal/v505/n7484/full/nature12912.html?WT.ec_id=NATURE-20140123
TCGA年度研讨会资料分享
TCGA想必搞生信都或有耳闻,尤其是癌症研究方向的,共4个年度研讨会,主要是pdf格式的ppt分享,有需要的可以具体点击到页面一个个下载自己慢慢研究,也可以用我下面链接直接下载。
本来是有youtube分享演讲视频的,但是国内被墙了,大家就看看ppt吧
http://www.genome.gov/17516564
The Cancer Genome Atlas (TCGA) is a comprehensive and coordinated effort to accelerate our understanding of the molecular basis of cancer through the application of genome analysis technologies, including large-scale genome sequencing.
TCGA is a joint effort of the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), which are both part of the National Institutes of Health, U.S. Department of Health and Human Services.
Meetings
- The Cancer Genome Atlas Fourth Annual Scientific Symposium

May 11-12, 2015 - The Cancer Genome Atlas Third Annual Scientific Symposium
May 12-13, 2014 - The Cancer Genome Atlas Second Annual Scientific Symposium
November 27-28, 2012 - The Cancer Genome Atlas First Annual Scientific Symposium
November 17-18, 2011
pdf链接地址如下
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Laird.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Durbin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Ley.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Sartor.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Ciriello.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Imielinski.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Gao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Carter.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Ng.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Parvin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Raphael.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Lawrence.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Kreisberg.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Marra.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Helman.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Stuart.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Cooper.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Levine.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Natsoulis.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Haussler.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Erkkila.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Gehlenborg.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Qiao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Sivachenko.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Sumazin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Gutman.pdf
http://www.genome.gov/Multimedia/Slides/TCGA1/TCGA1_Mardis.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/01_Shaw.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/02_Chanock.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/03_Staudt.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/05_Creighton.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/06_Stojanov.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/07_Karchin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/08_Mungall.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/09_Hakimi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/10_Gao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/11_Hayes.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/12_Troester.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/13_Knobluach.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/14_Raphael.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/15_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/16_Giordano.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/17_Weinstein.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/18_Zheng.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/19_Getz.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/20_VanDneBroek.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/21_Liao.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/22_Khazanov.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/23_Levine.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/24_Miller.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/25_Ewing.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/26_Cirello.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/27_Verhaak.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/28_Hofree.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/29_Meyerson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/30_Yang.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/31_Wheeler.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/32_Parfenov.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/33_Bernard-Rovira.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/34_Hast.pdf
http://www.genome.gov/Multimedia/Slides/TCGA2/36_Sellars.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/04_Brat.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/05_Mungall.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/06_Boutros.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/07_Zmuda.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/08_Benz.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/09_Zheng.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/11_Creighton.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/12_Aksoy.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/13_Dinh.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/14_Stuart.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/15_Amin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/16_Gross.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/15_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/18_Giordano.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/19_Amin-Mansour.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/20_Oesper.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/21_Gatza.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/22_Bernard.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/23_Sinha.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/24_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/25_Watson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/26_Martignetti.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/27_Bandlamudi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/28_Fu.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/29_Akdemir.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/30_Bass.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/31_Hakimi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/32_Wheeler.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/33_Lehmann.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/34_Gordenin.pdf
http://www.genome.gov/Multimedia/Slides/TCGA3/35_Wyczalkowski.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/02_Zenklusen.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/03_Hutter.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/04_Brat.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/05_Mungall.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/06_Linehan.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/07_Brooks.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/08_Wu.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/09_Giger.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/10_Wilkerson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/11_Orsulic.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/12_Zhong.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/13_Knijnenburg.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/14_Akbani.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/15_Wang.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/16_Poisson.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/17_Alaeimahabadi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/18_Noushmehr.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/19_Pantazi.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/20_Shih.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/21_Stransky.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/22_Giordano.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/23_Davidsen.pdf
http://www.genome.gov/Multimedia/Slides/TCGA4/24_Gross.pdf