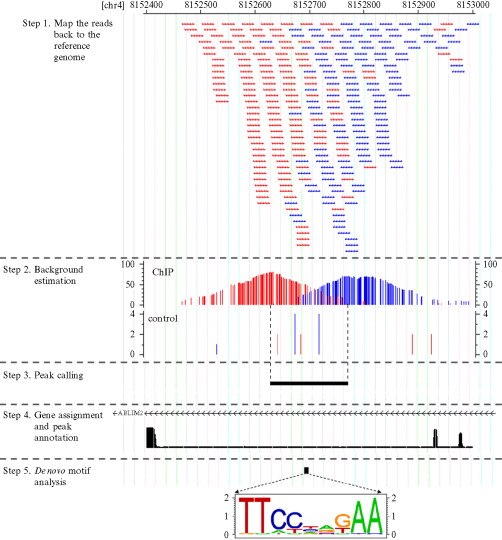
我只能说,CHIP-seq的确是非常完善的NGS流程了,各种资料层出不穷,大家首先可以看下面几个完整流程的PPT来对CHIP-seq流程有个大致的印象,我对前面提到的文献数据处理的几个要点,就跟下面这个图片类似:
QuEST is a statistical software for analysis of ChIP-Seq data with data and analysis results visualization through UCSC Genome Browser. http://www-hsc.usc.edu/~valouev/QuEST/QuEST.html
peak calling 阈值的选择: http://www.nature.com/nprot/journal/v7/n1/fig_tab/nprot.2011.420_F2.html
MeDIP-seq and histone modification ChIP-seq analysis http://crazyhottommy.blogspot.com/2014/01/medip-seq-and-histone-modification-chip.html
2011-review-CHIP-seq-high-quaility-data: http://www.nature.com/ni/journal/v12/n10/full/ni.2117.html?message-global=remove
不同处理条件的CHIP-seq的差异peaks分析: http://www.slideshare.net/thefacultyl/diffreps-automated-chipseq-differential-analysis-package
一个实际的CHIP-seq数据分析例子: http://www.biologie.ens.fr/~mthomas/other/chip-seq-training/