Lecture3_One-hundred-software-in-bioinformatics094_ConsensusClusterPlus
Jimmy(jmzeng1314@outlook.com)
Keywords: ConsensusClusterPlus/clust/R/package
Data: ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE62nnn/GSE62254/matrix/GSE62254_series_matrix.txt.gz
#source("http://bioconductor.org/biocLite.R")
#biocLite("ConsensusClusterPlus")
library(ConsensusClusterPlus)
#wget ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE62nnn/GSE62254/matrix/GSE62254_series_matrix.txt.gz
a=read.table("GSE62254_series_matrix.txt.gz",comment.char = "!",stringsAsFactors=F,header=T)
rownames(a)=a[,1]
a=a[,-1]
#> dim(a)
#[1] 54675 300
d=a
mads=apply(d,1,mad)
d=d[rev(order(mads))[1:5000],]
d = sweep(d,1, apply(d,1,median,na.rm=T))
title="./"
#所有的图片以及数据都会输出到这里的
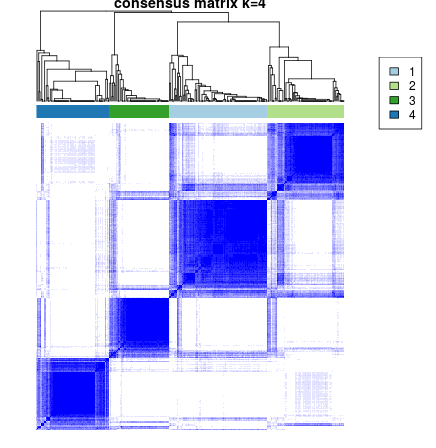
results = ConsensusClusterPlus(as.matrix(d),maxK=6,reps=50,pItem=0.8,pFeature=1,
title=title,clusterAlg="hc",distance="pearson",seed=1262118388.71279,plot="png")
Subtype: 4
useful links : http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62254
http://www.bio-info-trainee.com/?p=945
http://www.bio-info-trainee.com/?p=969
http://www.bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.html